Relational data: a quick review
Relational data is multiple tables of data that when combined together answer research questions. Relations define the important element, not just the individual tables. Relations are defined between a pair of tables, or potentially complex structures can be built up with more than 2 tables. In many situations, data is stored in a relational format because to do otherwise would introduce redundancy and use unnecessary storage space.
This data structure requires relational verbs to combine data across tables. Mutating joins add new variables to one data frame from matching observations in another, whereas filtering joins filter observations from one data frame based on whether or not they match an observation in the other table.
superheroes and publishers
Let’s review how these different types of joining operations work with relational data on comic books. Load the rcis library. There are two data frames which contain data on comic books.
library(tidyverse)
library(rcis)
superheroes
## # A tibble: 3 × 4
## name alignment gender publisher
## <chr> <chr> <chr> <chr>
## 1 Magneto bad male Marvel
## 2 Batman good male DC
## 3 Sabrina good female Archie Comics
publishers
## # A tibble: 3 × 2
## publisher yr_founded
## <chr> <dbl>
## 1 DC 1934
## 2 Marvel 1939
## 3 Image 1992
Would it make sense to store these two data frames in the same tibble? No! This is because each data frame contains substantively different information:
superheroescontains data on superheroespublisherscontains data on publishers
The units of analysis are completely different. Just as it made sense to split Minard’s data into two separate data frames, it also makes sense to store them separately here. That said, depending on the type of analysis you seek to perform, it makes sense to join the data frames together temporarily. How should we join them? Well it depends on how you plan to ask your question. Let’s look at the result of several different join operations.
Mutating joins
Inner join
inner_join(x, y): Return all rows from x where there are matching values in y, and all columns from x and y. If there are multiple matches between x and y, all combination of the matches are returned. This is a mutating join.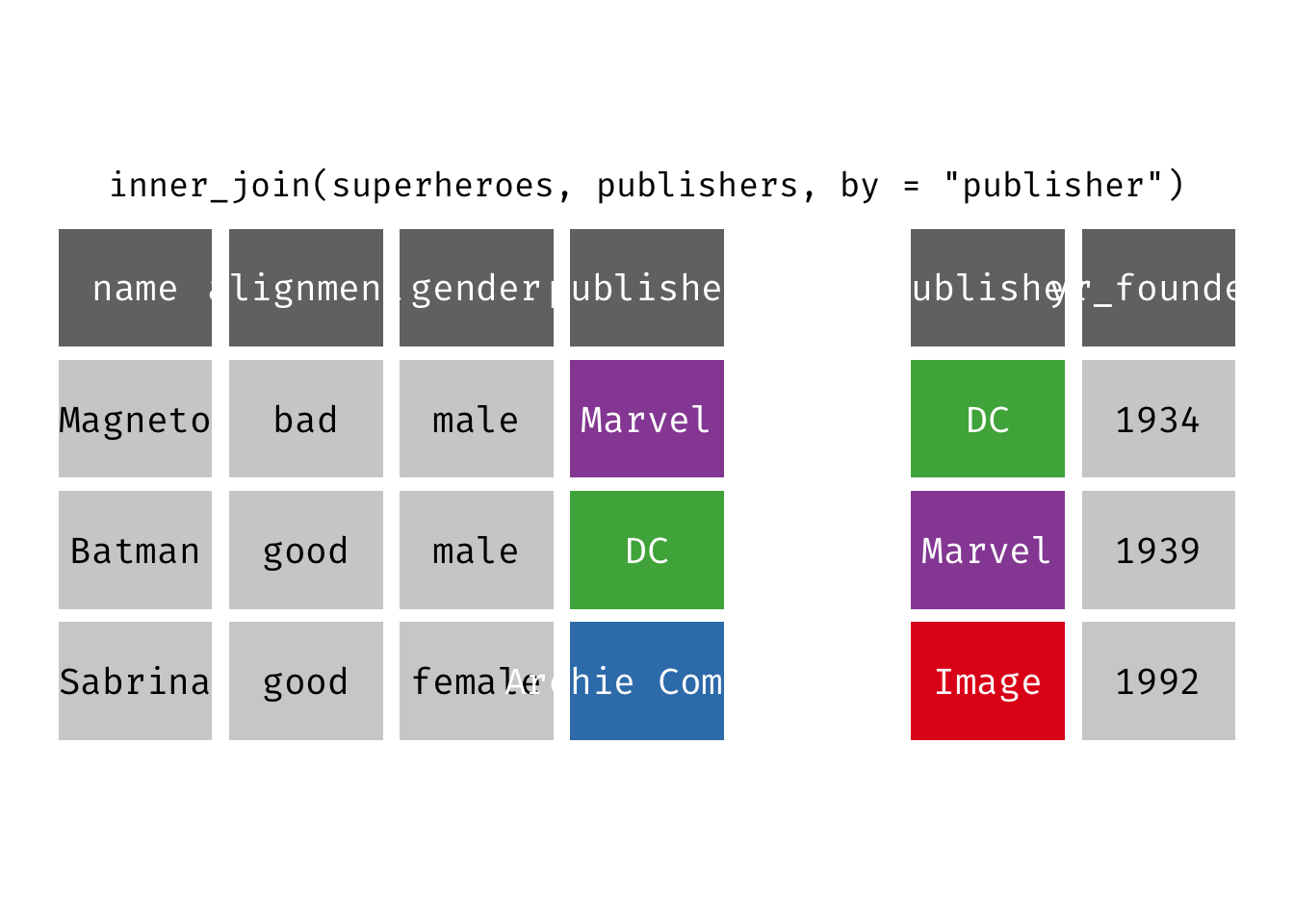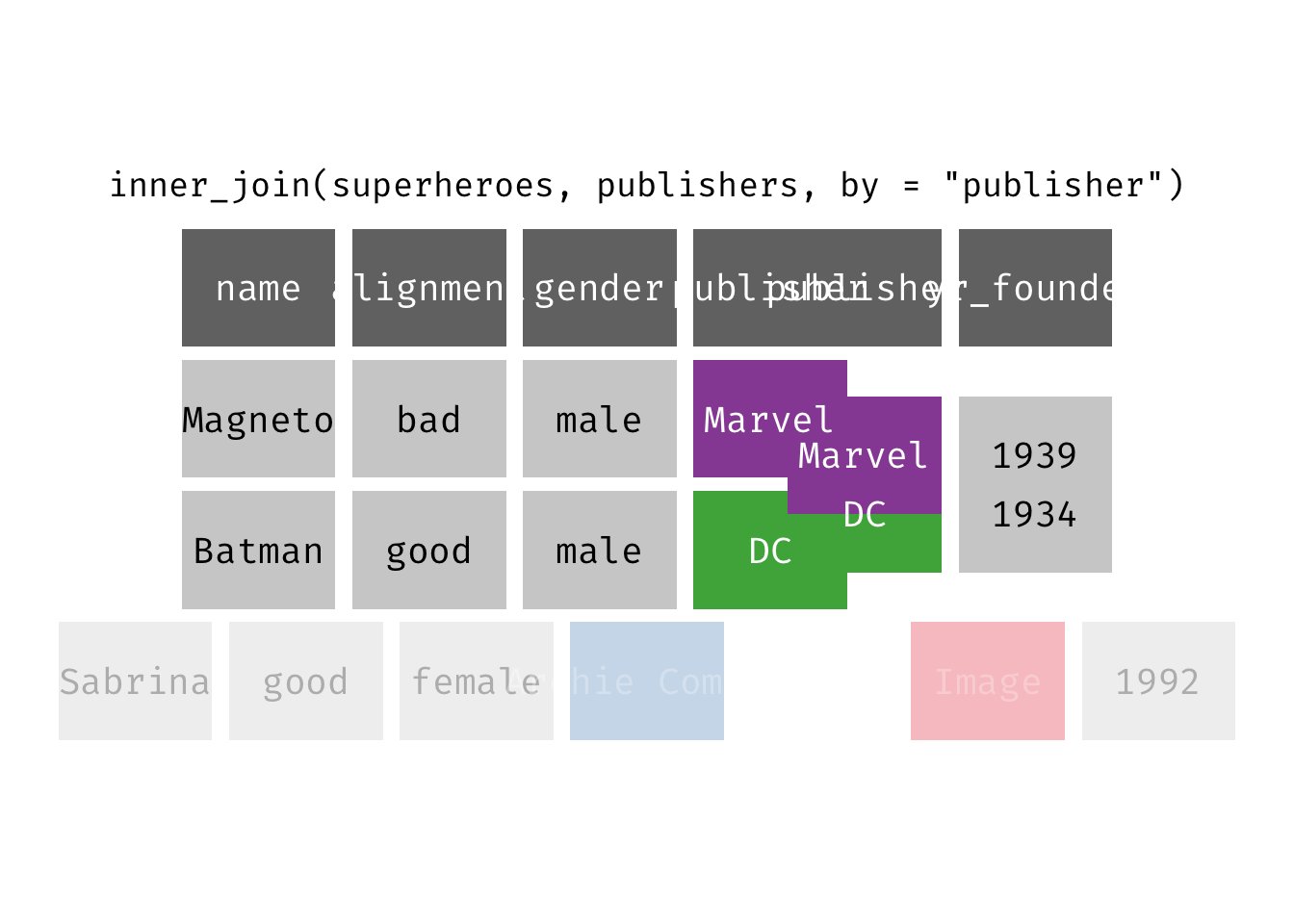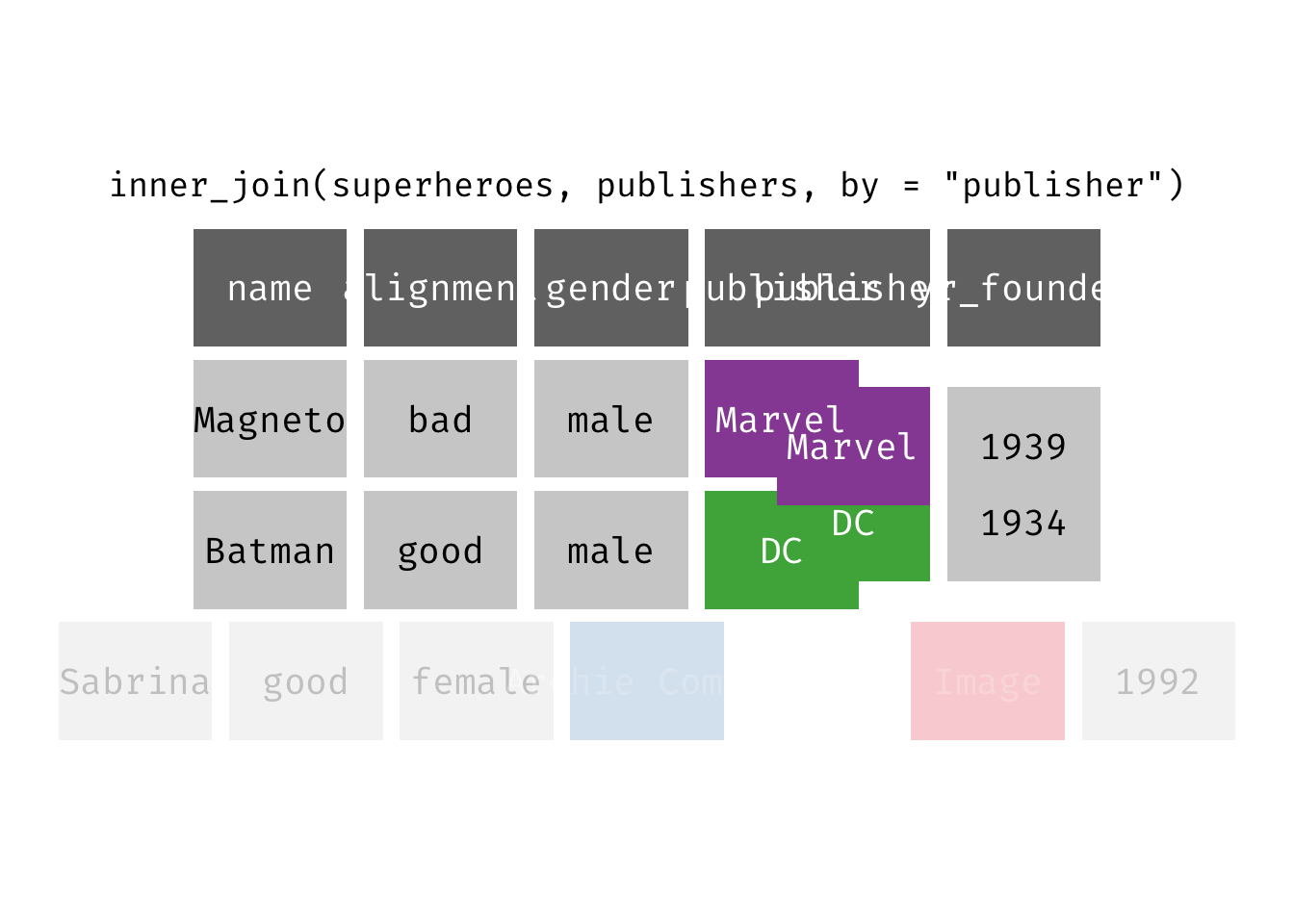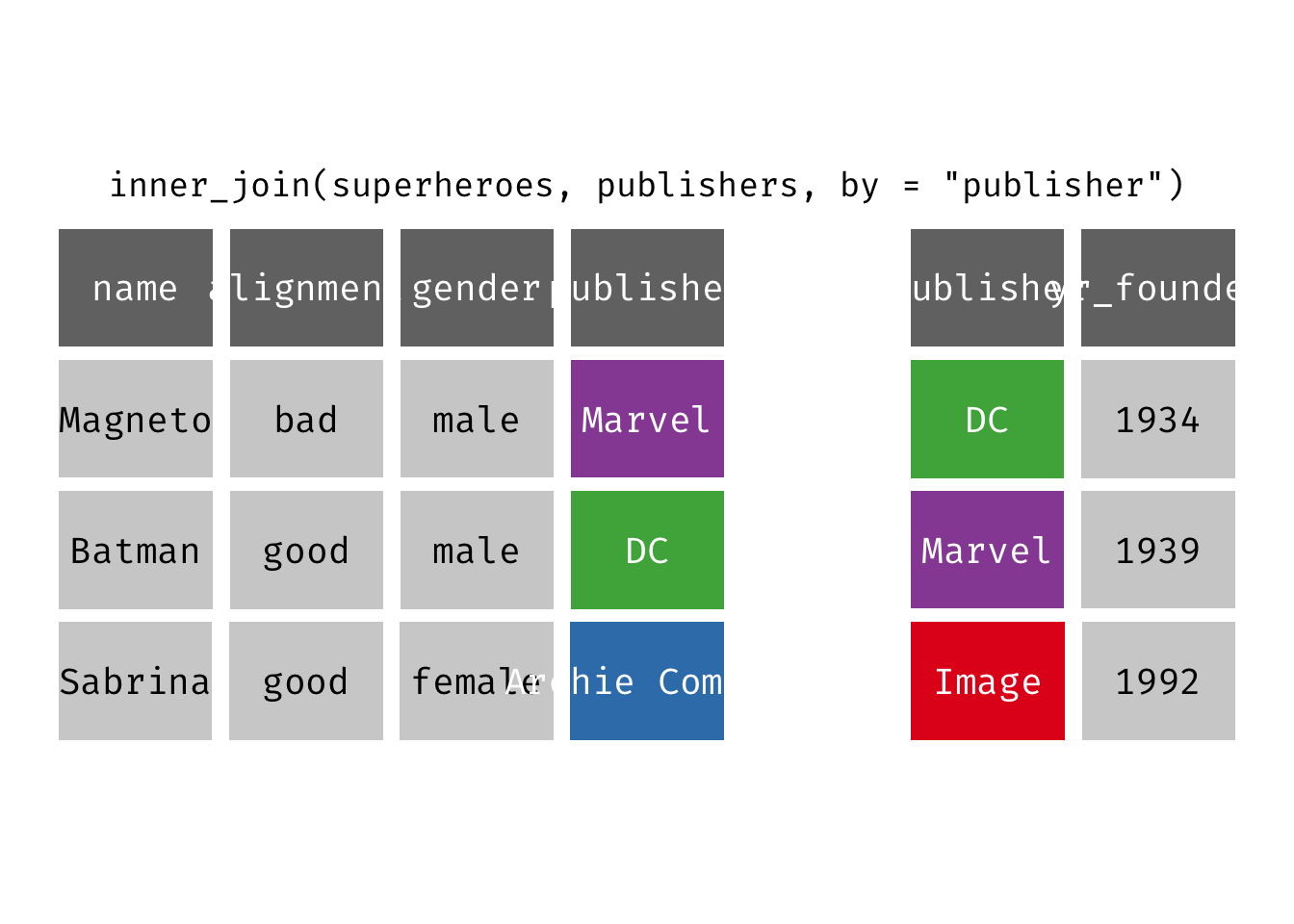
(ijsp <- inner_join(x = superheroes, y = publishers))
## Joining, by = "publisher"
## # A tibble: 2 × 5
## name alignment gender publisher yr_founded
## <chr> <chr> <chr> <chr> <dbl>
## 1 Magneto bad male Marvel 1939
## 2 Batman good male DC 1934
We lose Sabrina in the join because, although she appears in x = superheroes, her publisher Archie Comics does not appear in y = publishers. The join result has all variables from x = superheroes plus yr_founded, from y.
Left join
left_join(x, y): Return all rows from x, and all columns from x and y. If there are multiple matches between x and y, all combination of the matches are returned. This is a mutating join.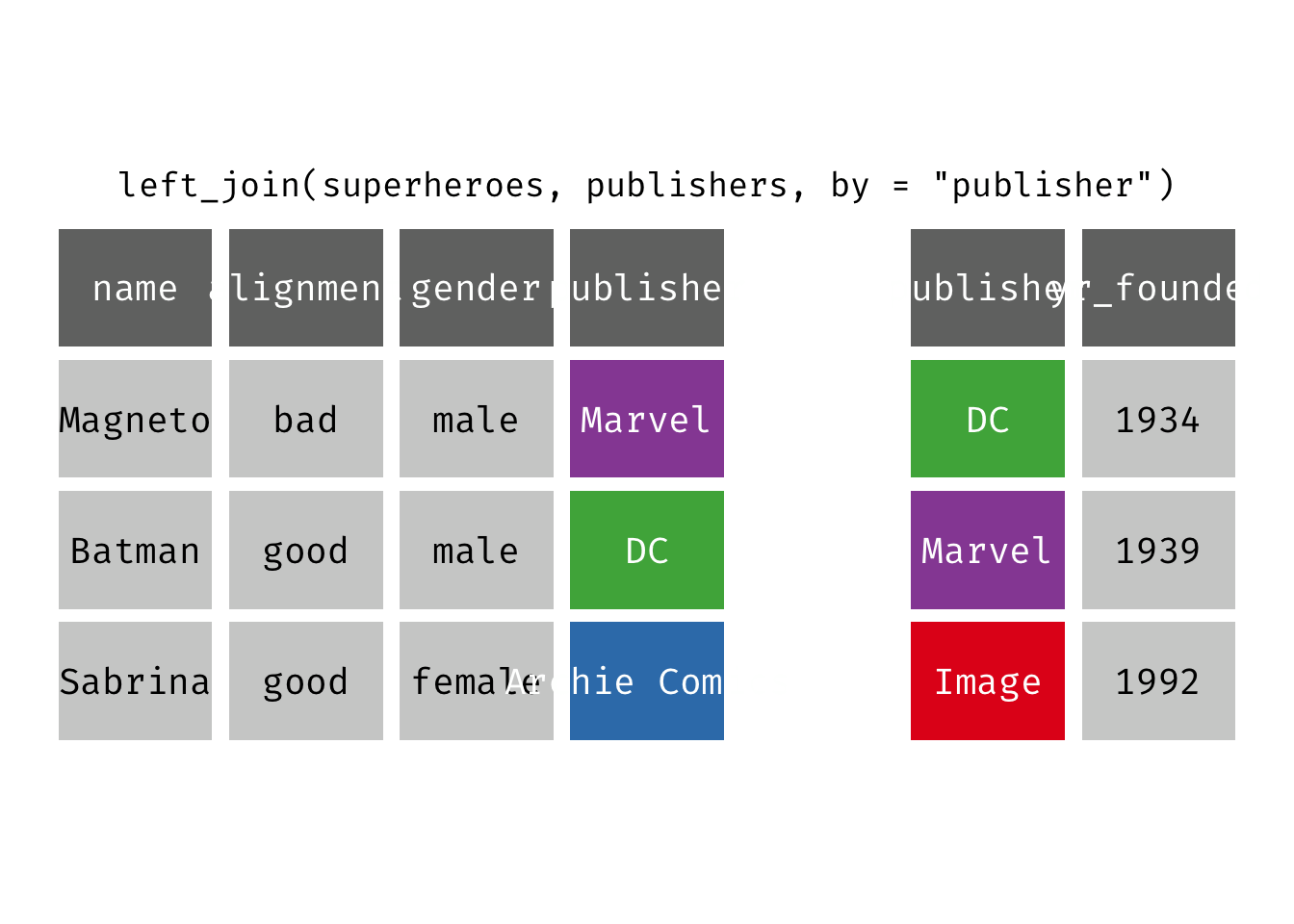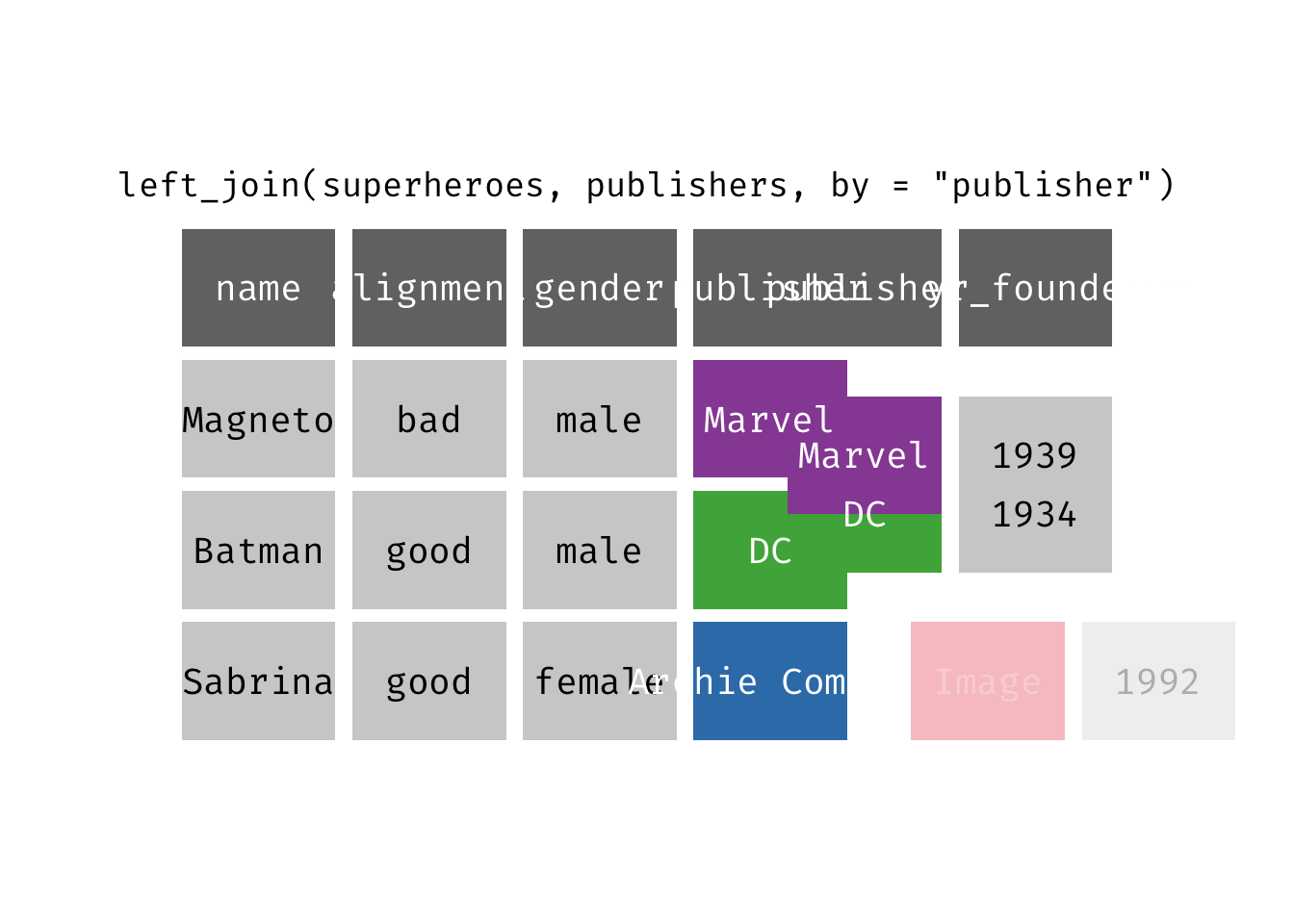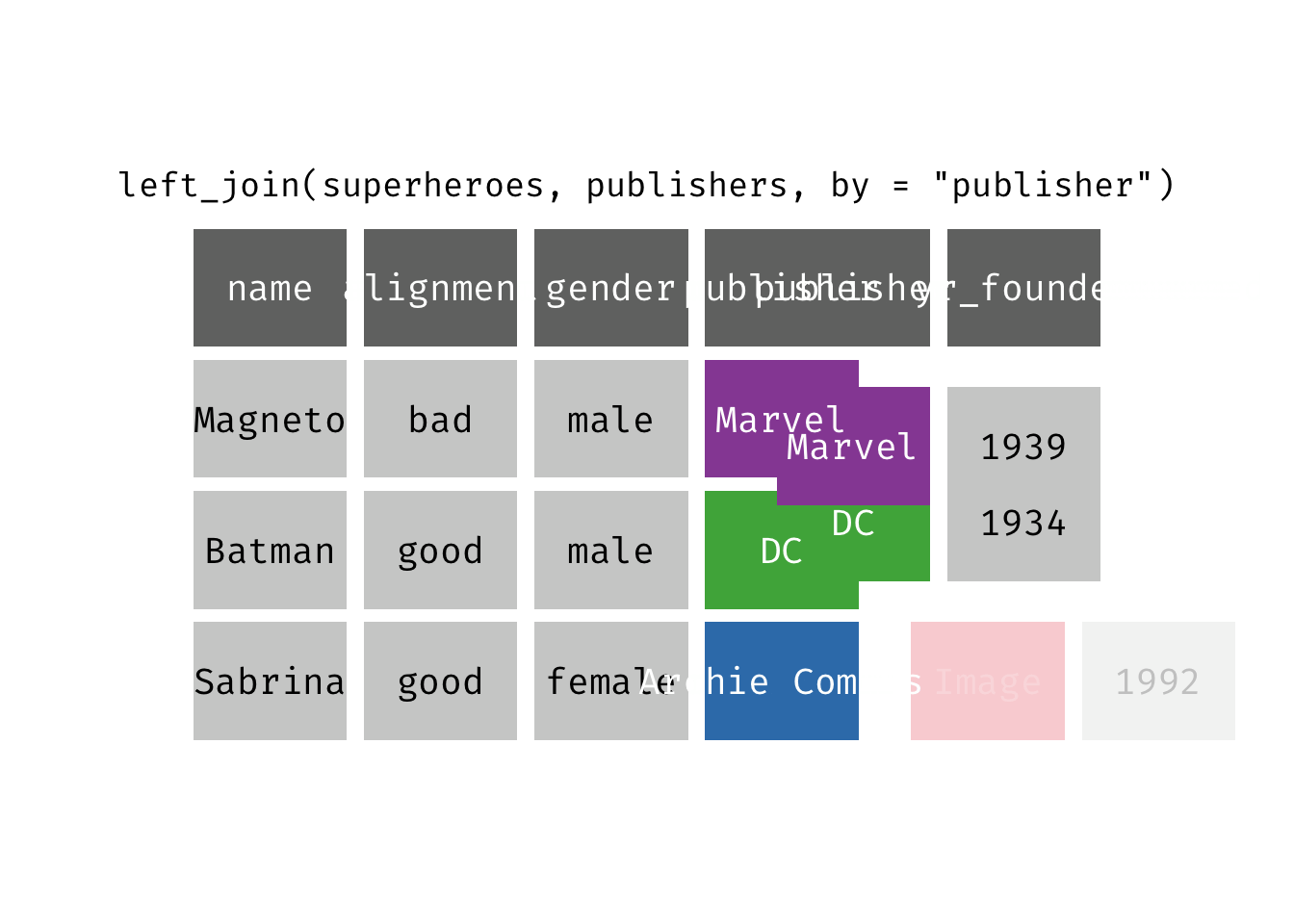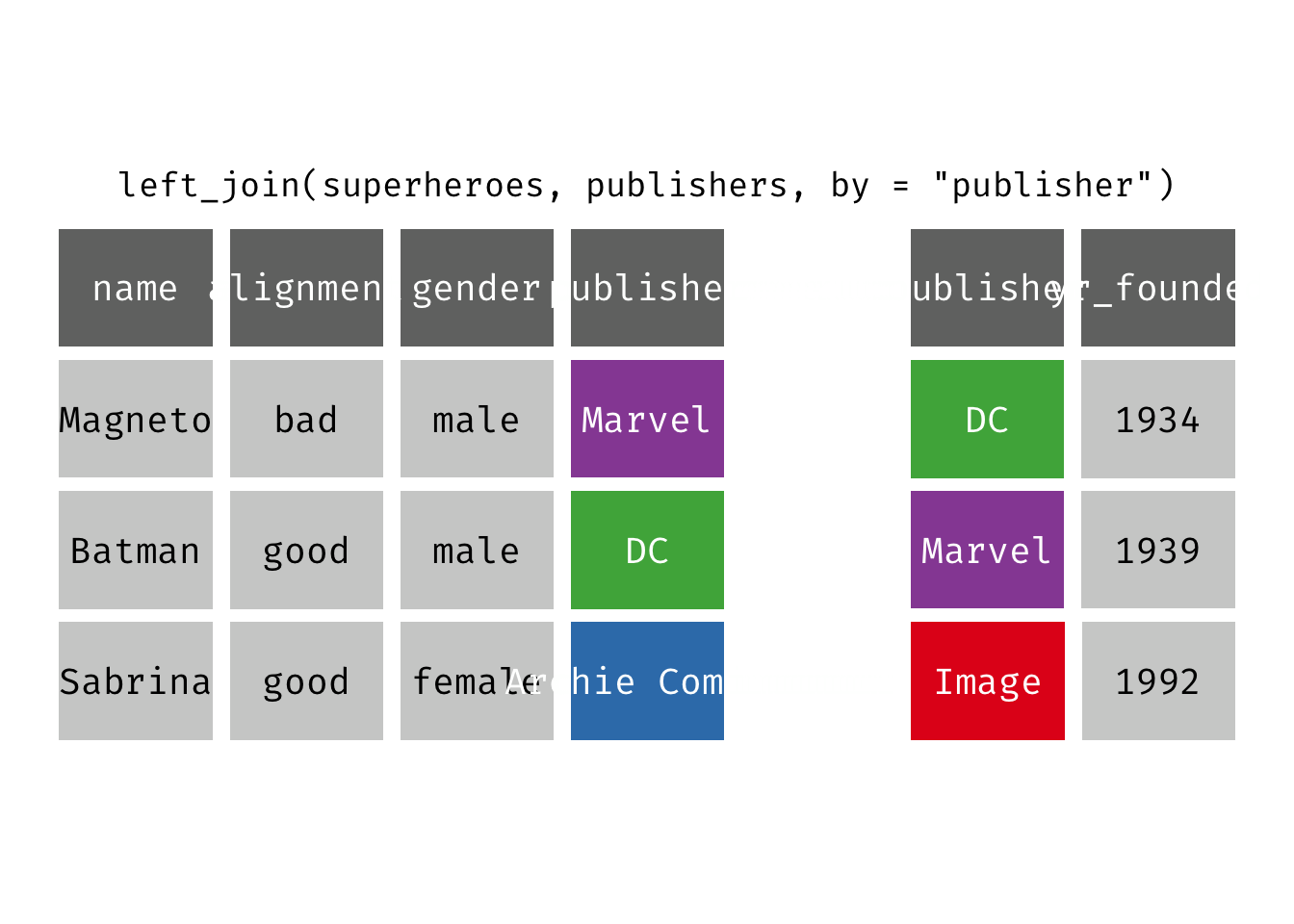
(ljsp <- left_join(x = superheroes, y = publishers))
## Joining, by = "publisher"
## # A tibble: 3 × 5
## name alignment gender publisher yr_founded
## <chr> <chr> <chr> <chr> <dbl>
## 1 Magneto bad male Marvel 1939
## 2 Batman good male DC 1934
## 3 Sabrina good female Archie Comics NA
We basically get x = superheroes back, but with the addition of variable yr_founded, which is unique to y = publishers. Sabrina, whose publisher does not appear in y = publishers, has an NA for yr_founded.
Right join
right_join(x, y): Return all rows from y, and all columns from x and y. If there are multiple matches between x and y, all combination of the matches are returned. This is a mutating join.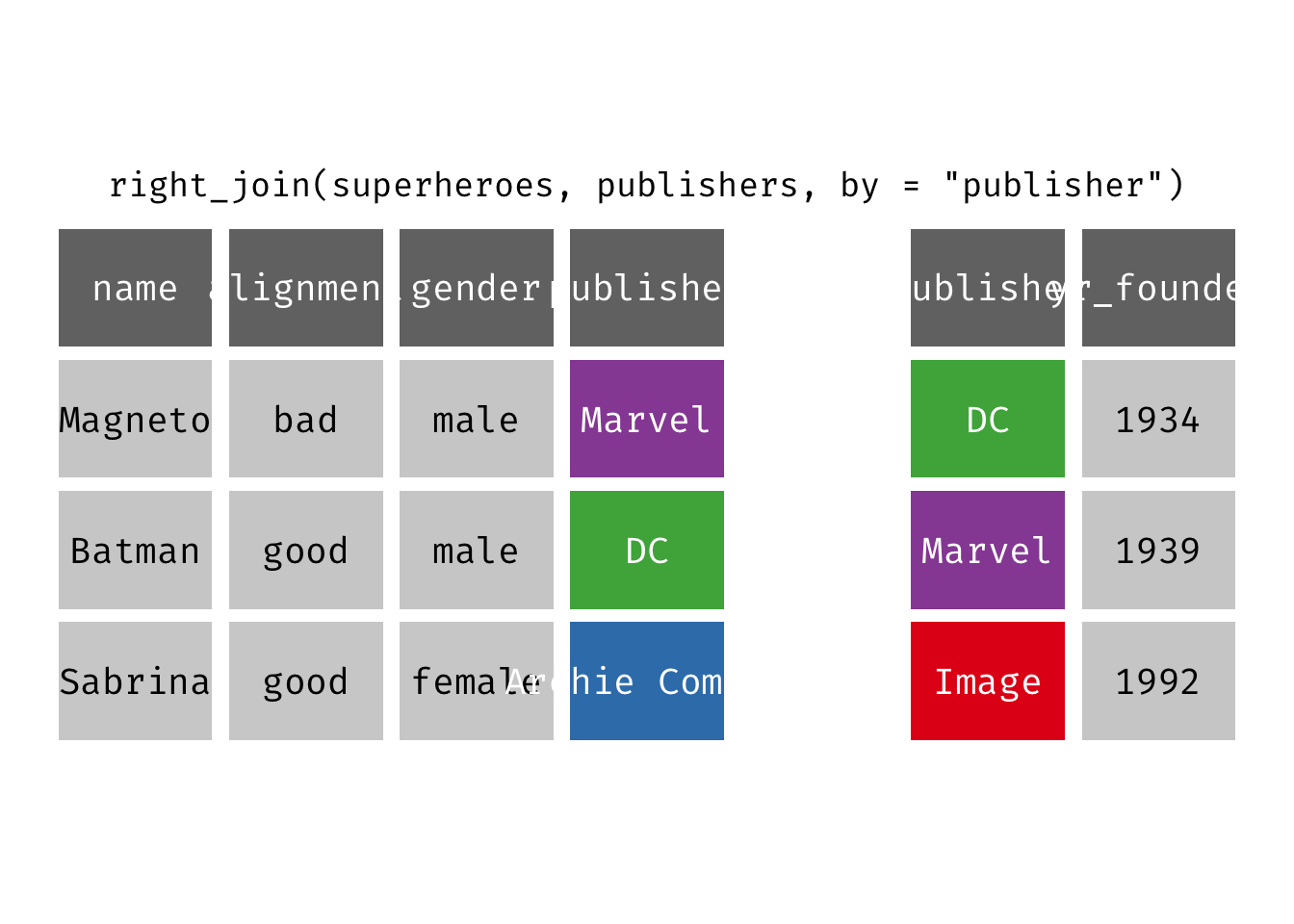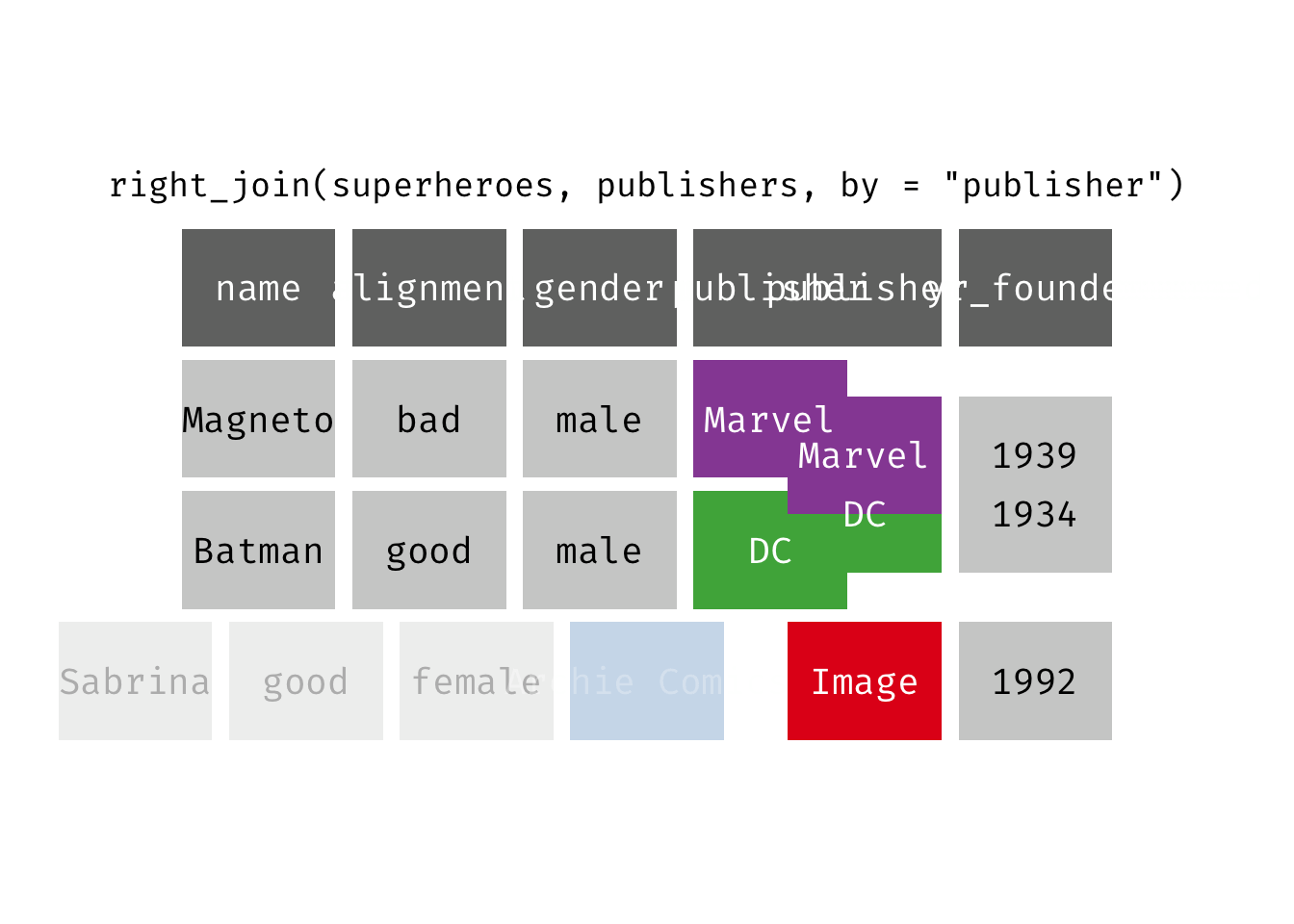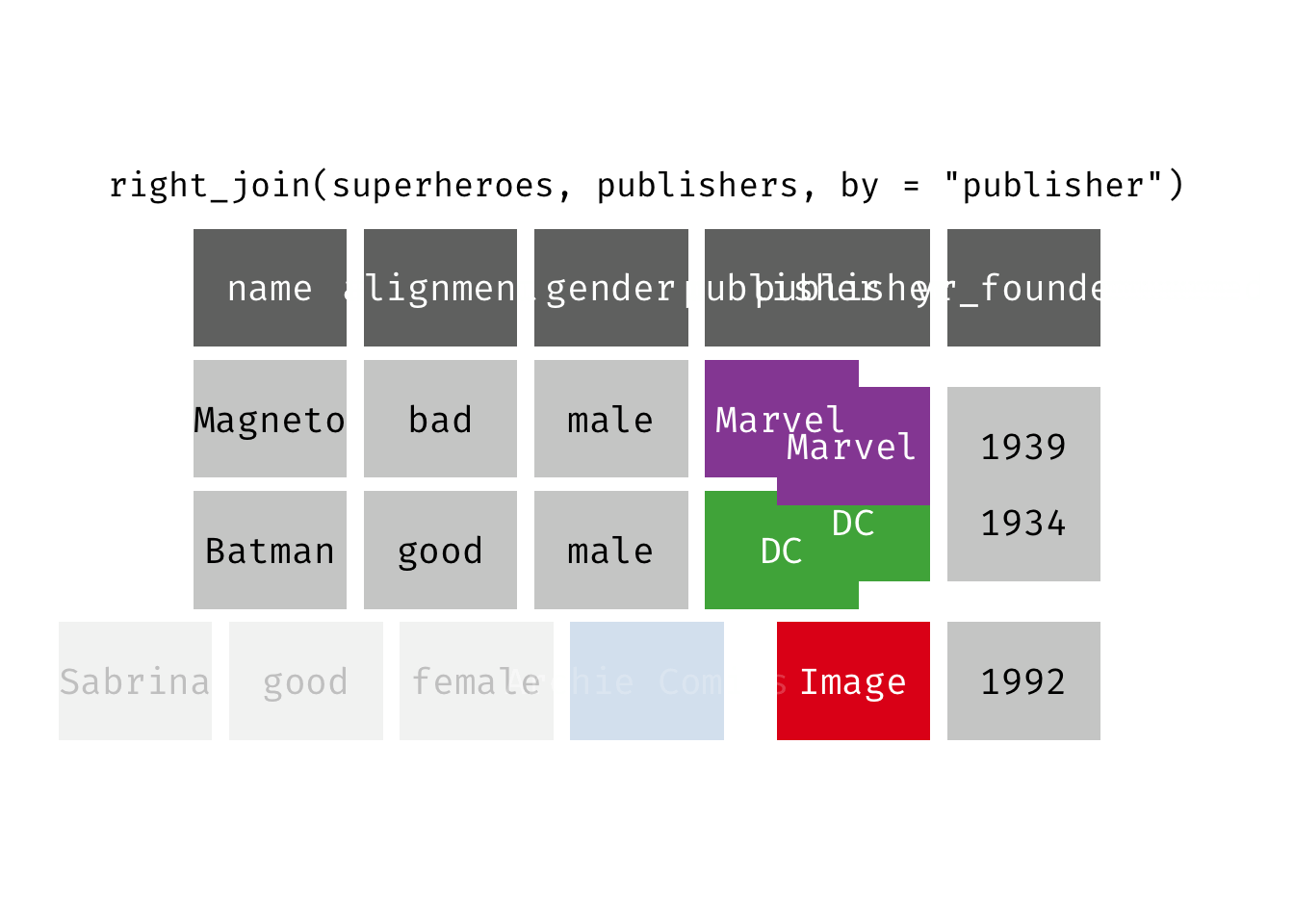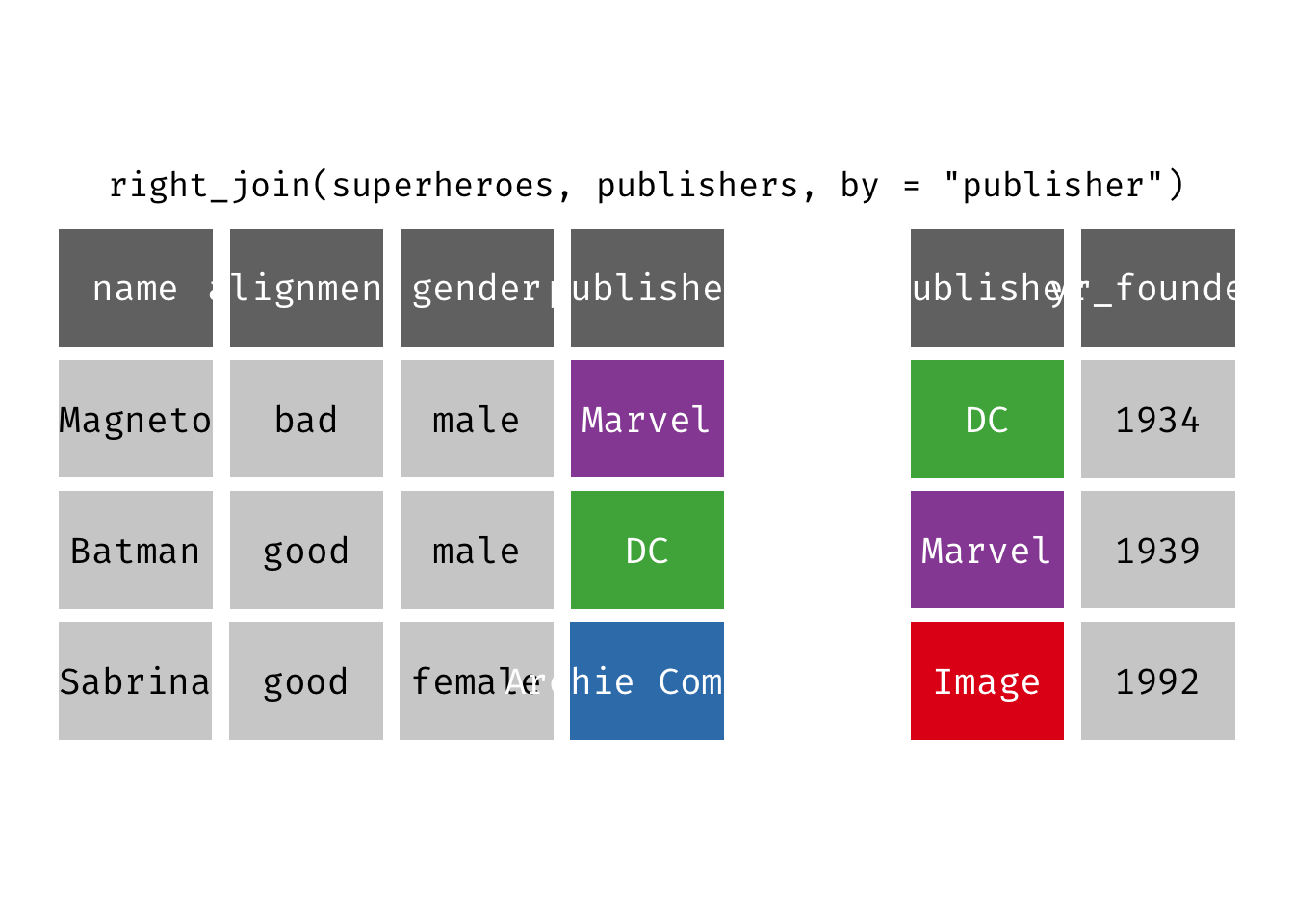
We basically get y = publishers back, but with the addition of variables name, alignment, and gender, which is unique to x = superheroes. Image, who did not publish any of the characters in superheroes, has an NA for the new variables.
We could also accomplish virtually the same thing using left_join() by reversing the order of the data frames in the function:
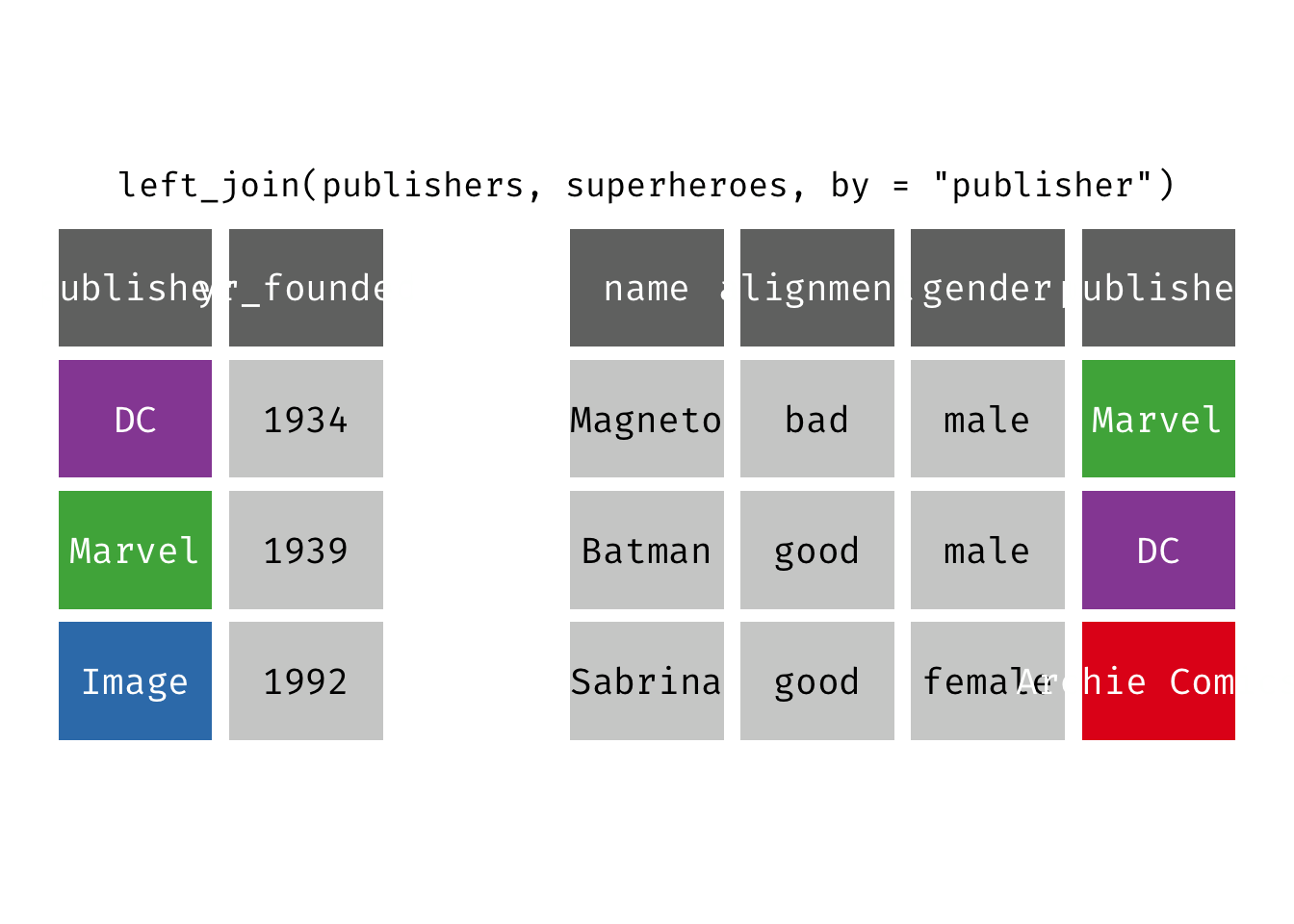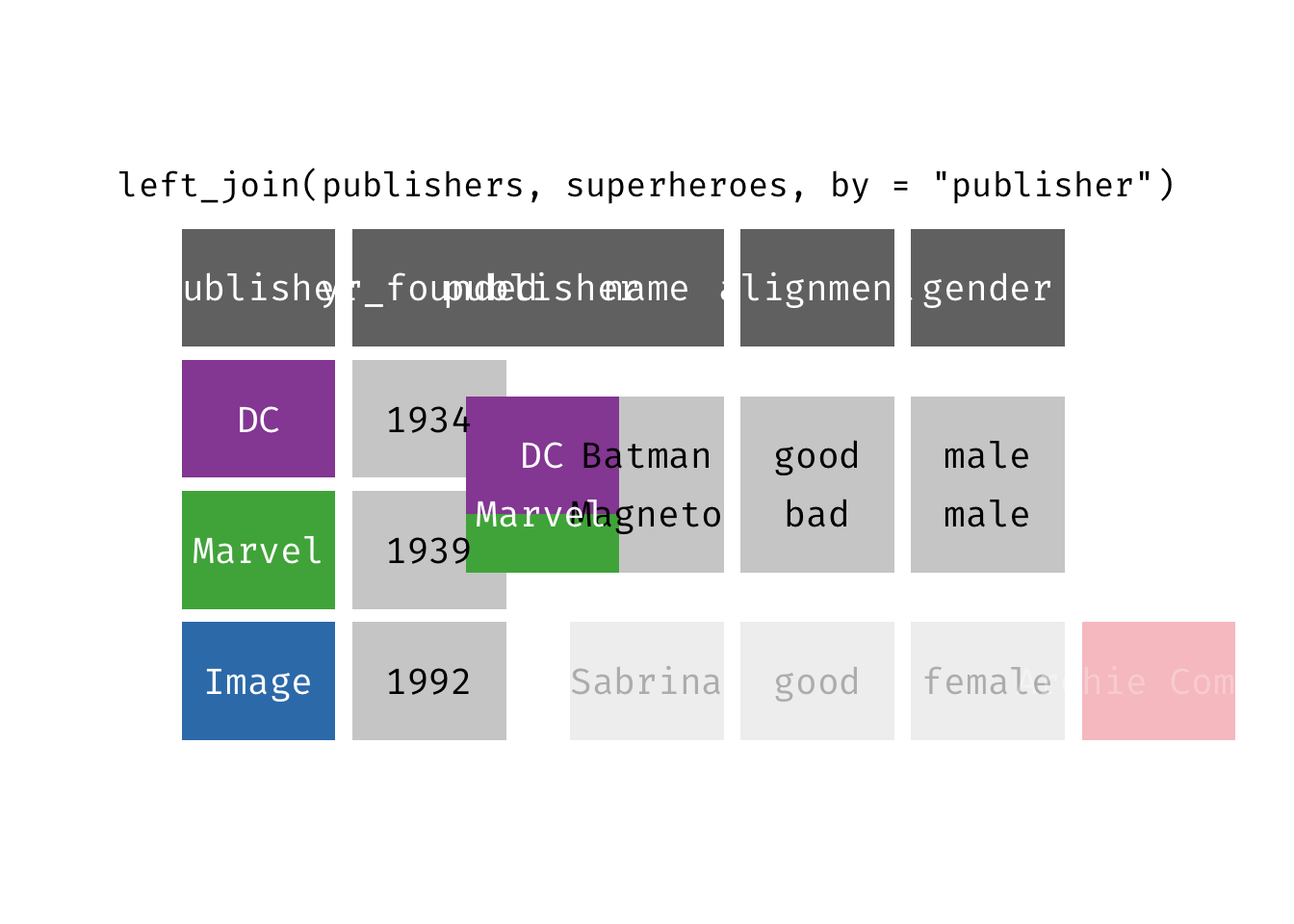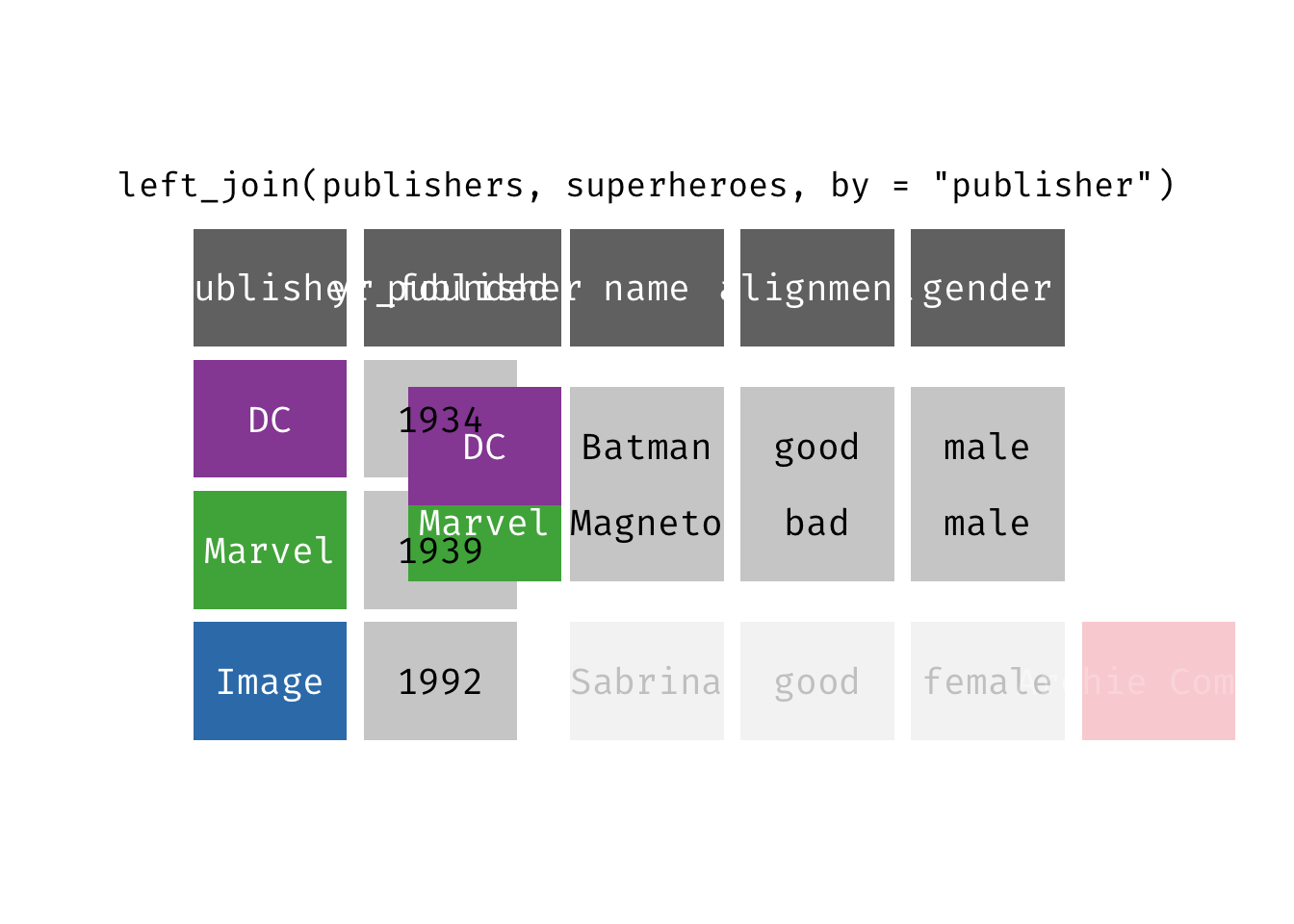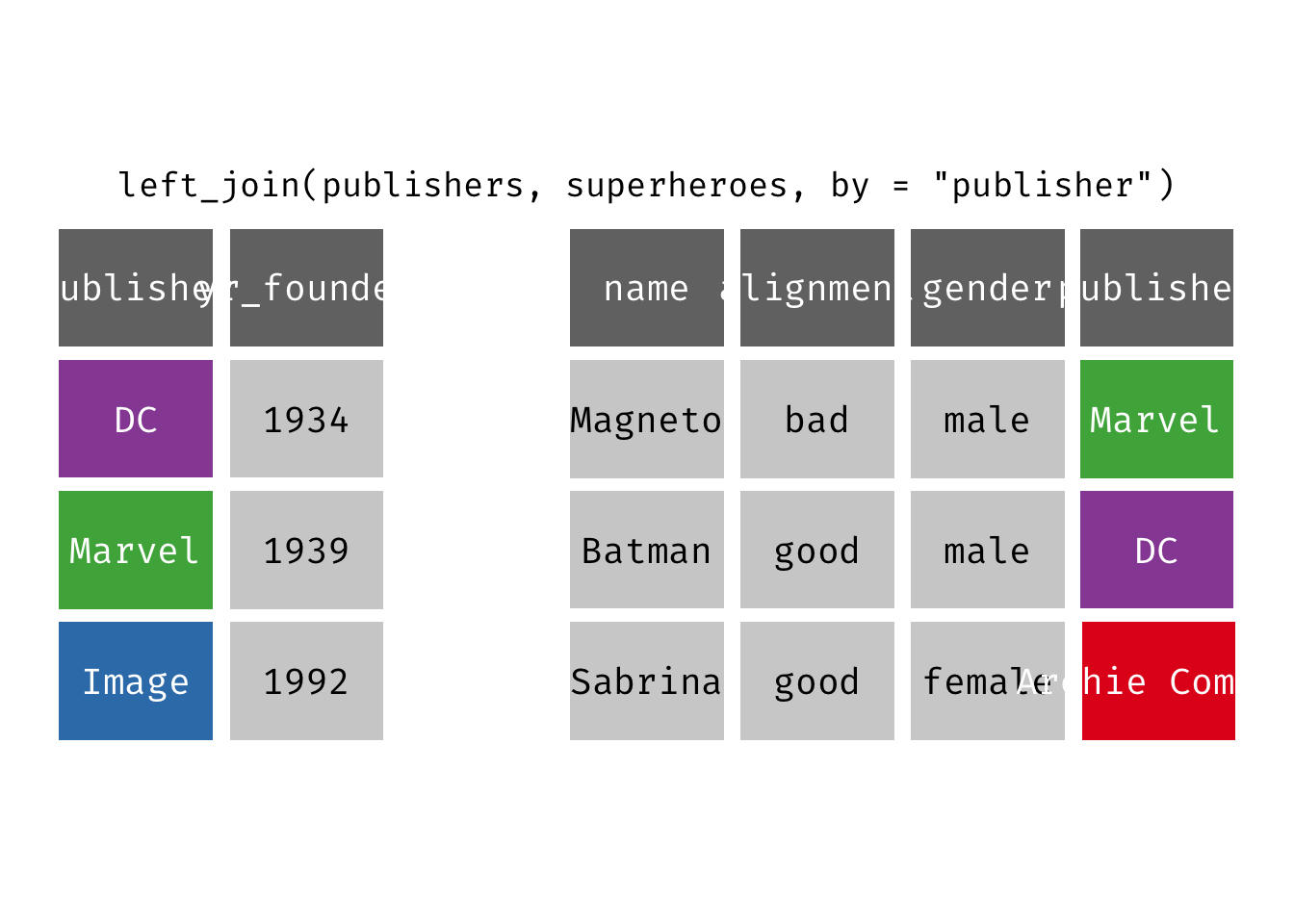
left_join(x = superheroes, y = publishers)
## Joining, by = "publisher"
## # A tibble: 3 × 5
## name alignment gender publisher yr_founded
## <chr> <chr> <chr> <chr> <dbl>
## 1 Magneto bad male Marvel 1939
## 2 Batman good male DC 1934
## 3 Sabrina good female Archie Comics NA
Doing so returns the same basic data frame, with the column orders reversed. right_join() is not used as commonly as left_join(), but works well in a piped operation when you perform several functions on x but then want to join it with y and only keep rows that appear in y.
Full join
full_join(x, y): Return all rows and all columns from both x and y. Where there are not matching values, returns NA for the one missing. This is a mutating join.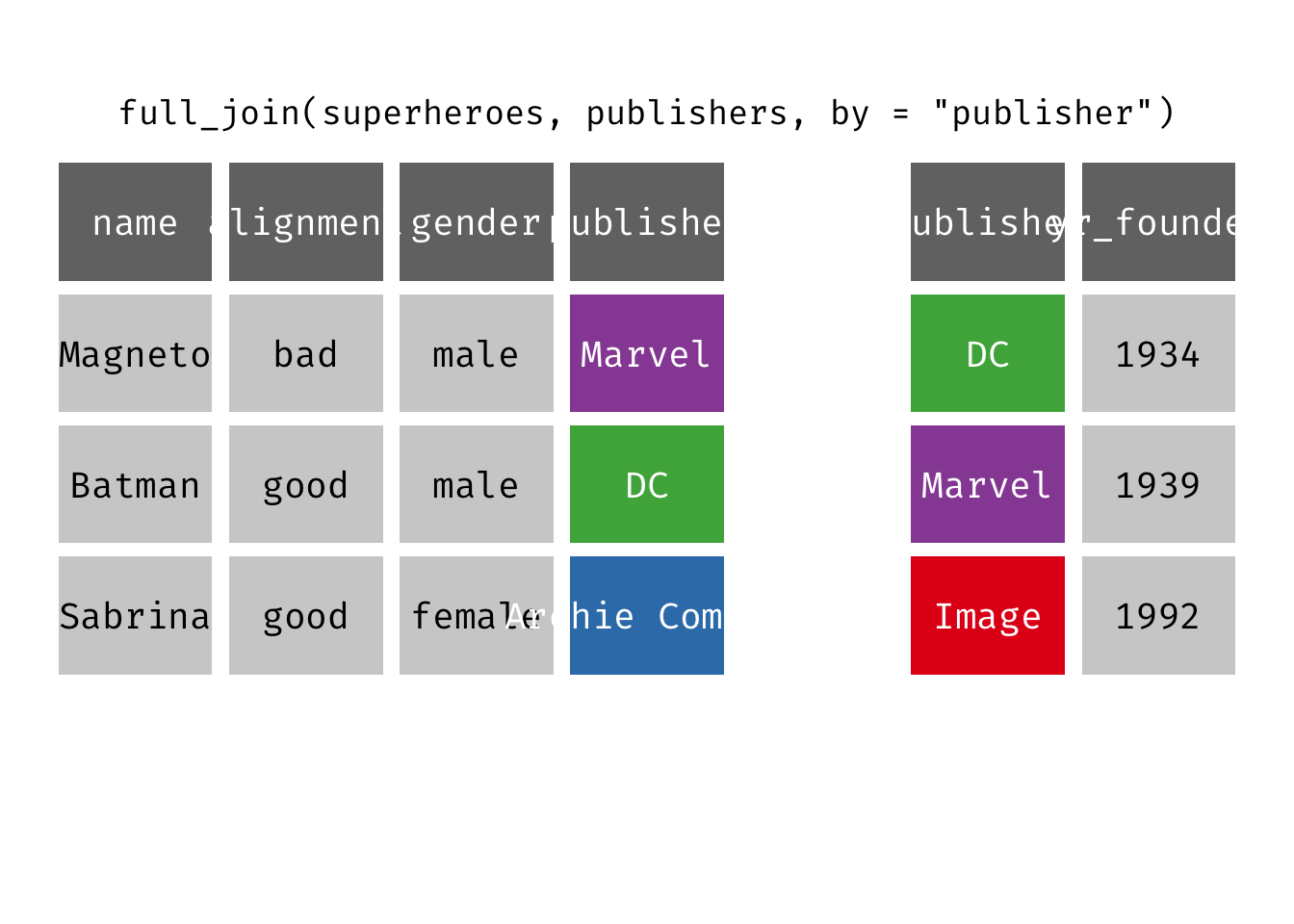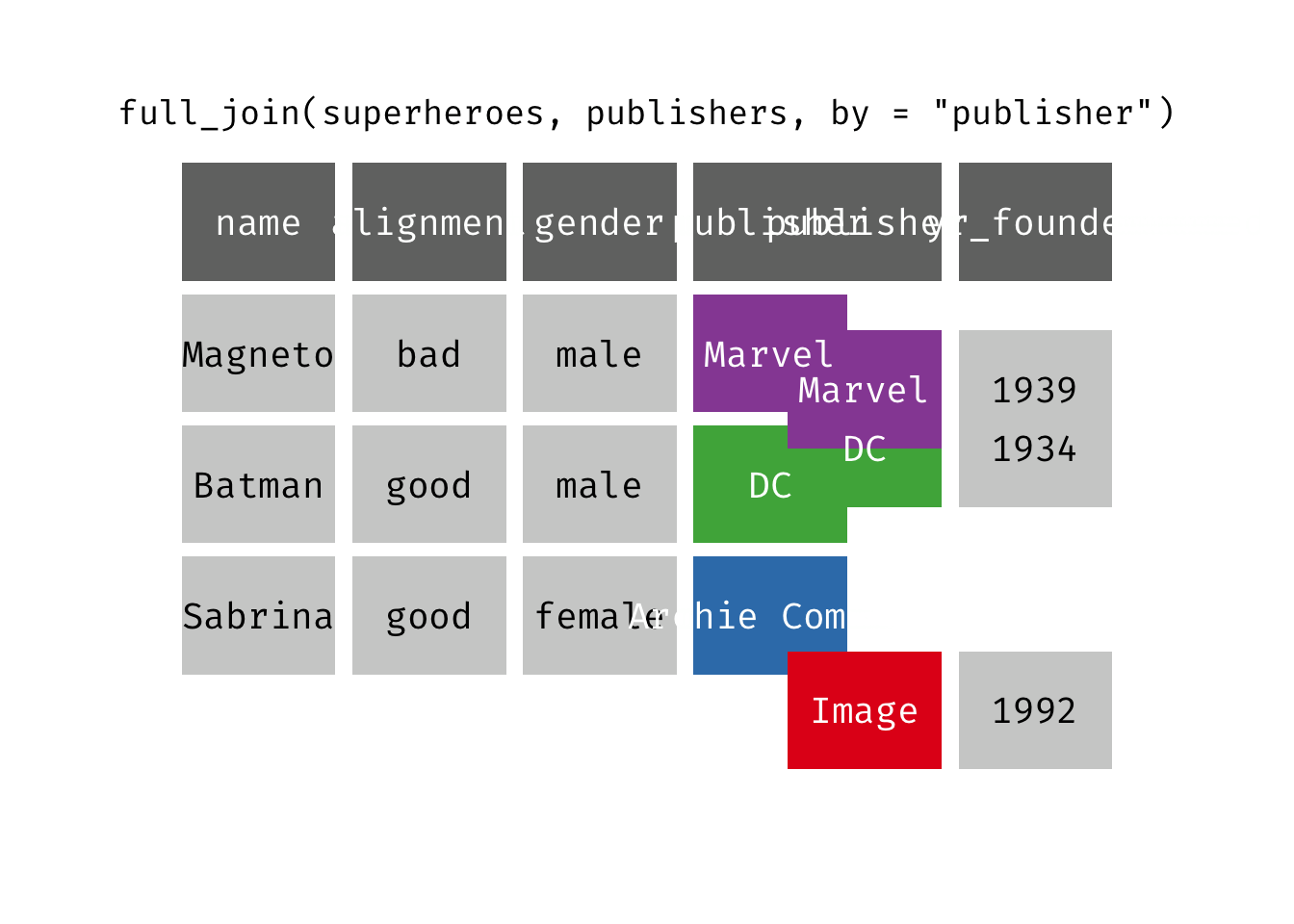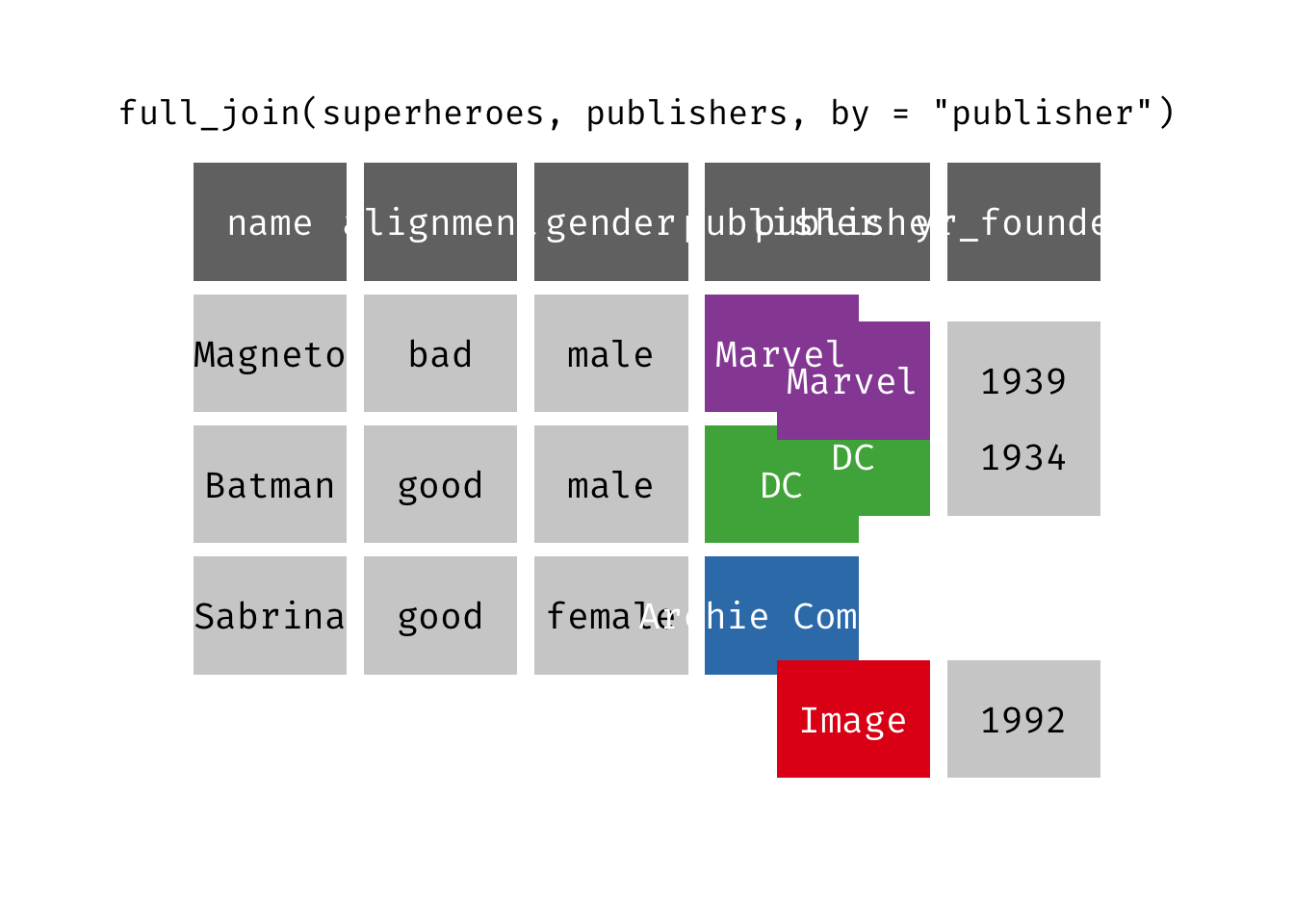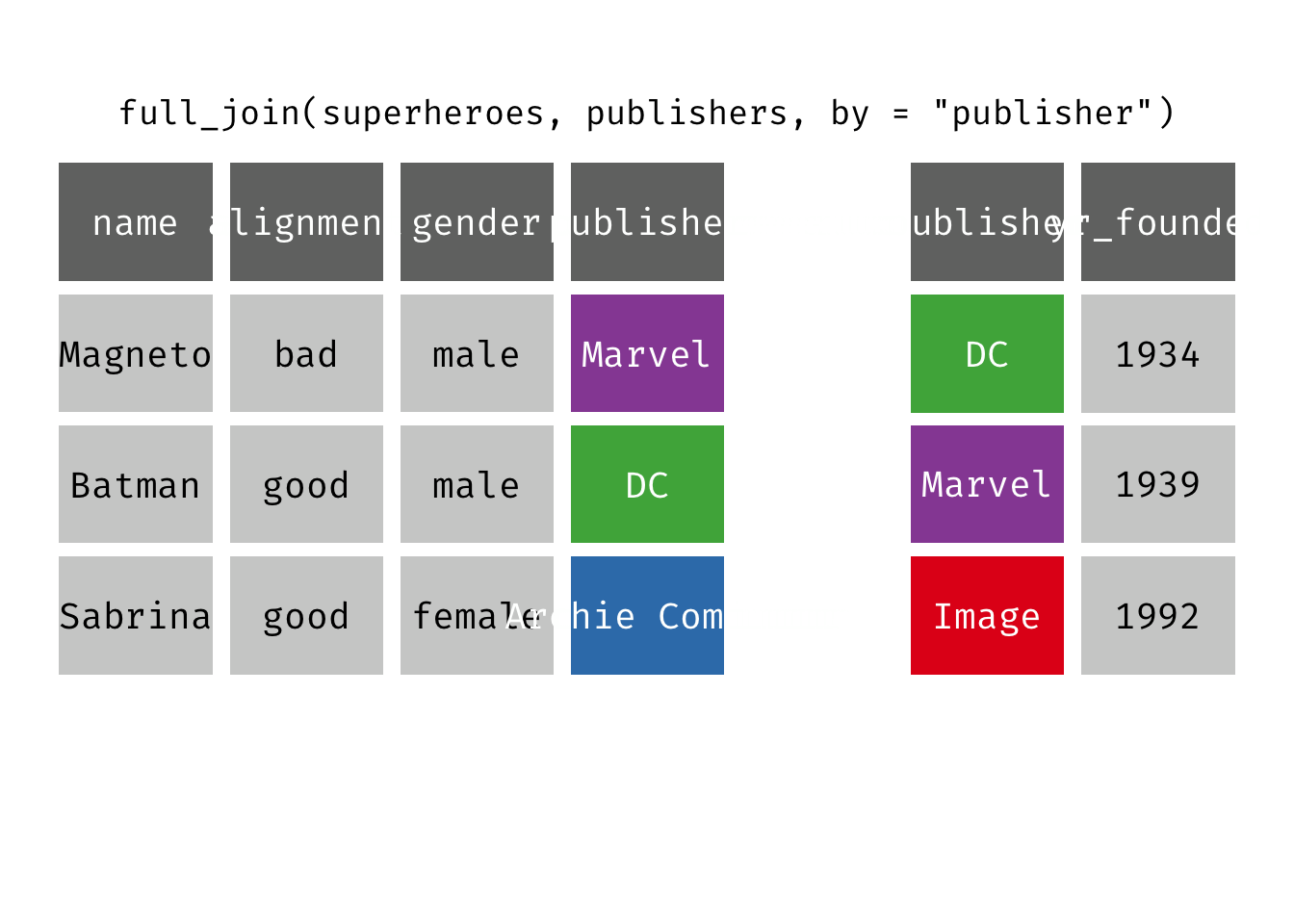
(fjsp <- full_join(x = superheroes, y = publishers))
## Joining, by = "publisher"
## # A tibble: 4 × 5
## name alignment gender publisher yr_founded
## <chr> <chr> <chr> <chr> <dbl>
## 1 Magneto bad male Marvel 1939
## 2 Batman good male DC 1934
## 3 Sabrina good female Archie Comics NA
## 4 <NA> <NA> <NA> Image 1992
We get all rows of x = superheroes plus a new row from y = publishers, containing the publisher “Image”. We get all variables from x = superheroes AND all variables from y = publishers. Any row that derives solely from one table or the other carries NAs in the variables found only in the other table.
Filtering joins
Semi join
semi_join(x, y): Return all rows from x where there are matching values in y, keeping just columns from x. A semi join differs from an inner join because an inner join will return one row of x for each matching row of y (potentially duplicating rows in x), whereas a semi join will never duplicate rows of x. This is a filtering join.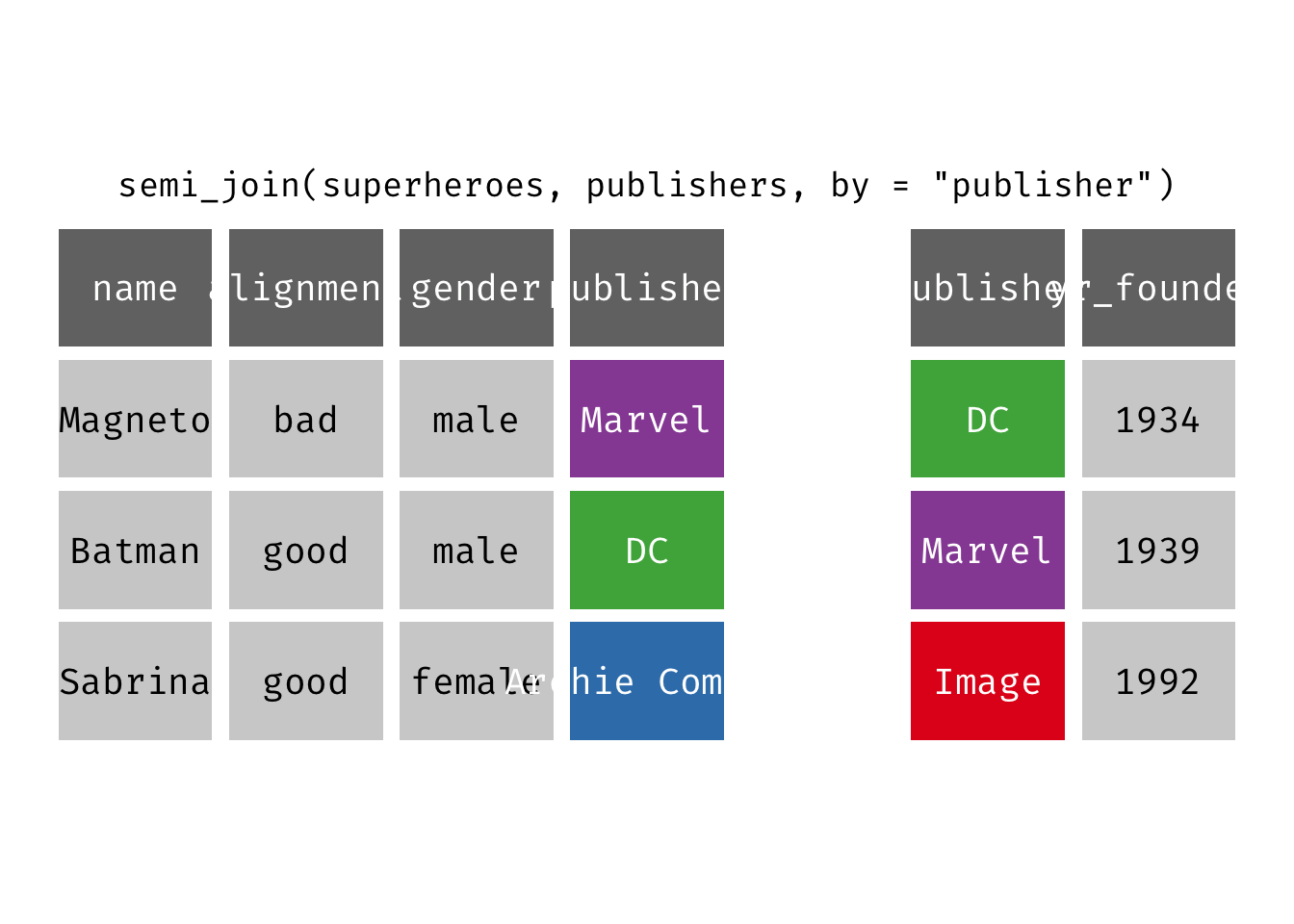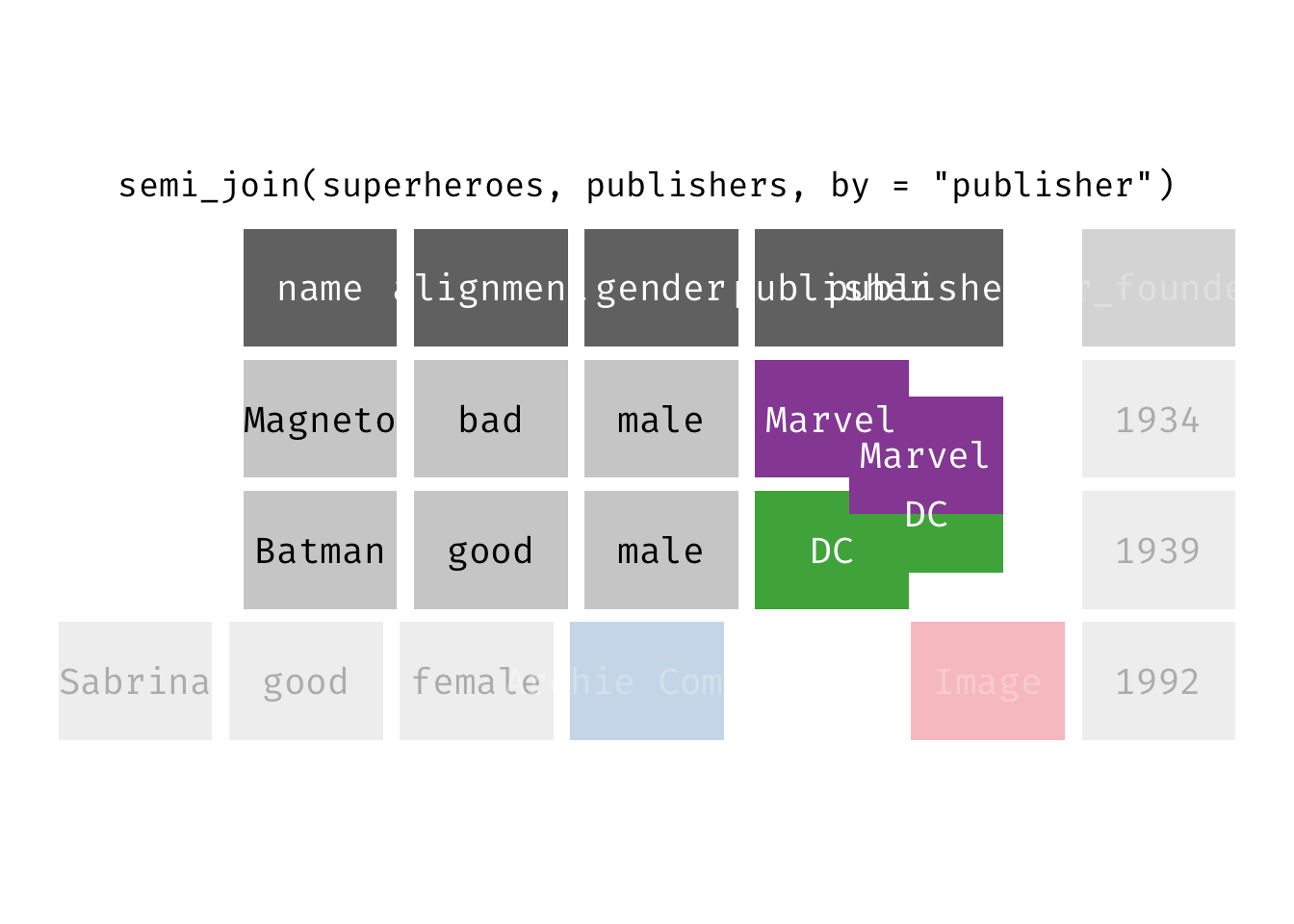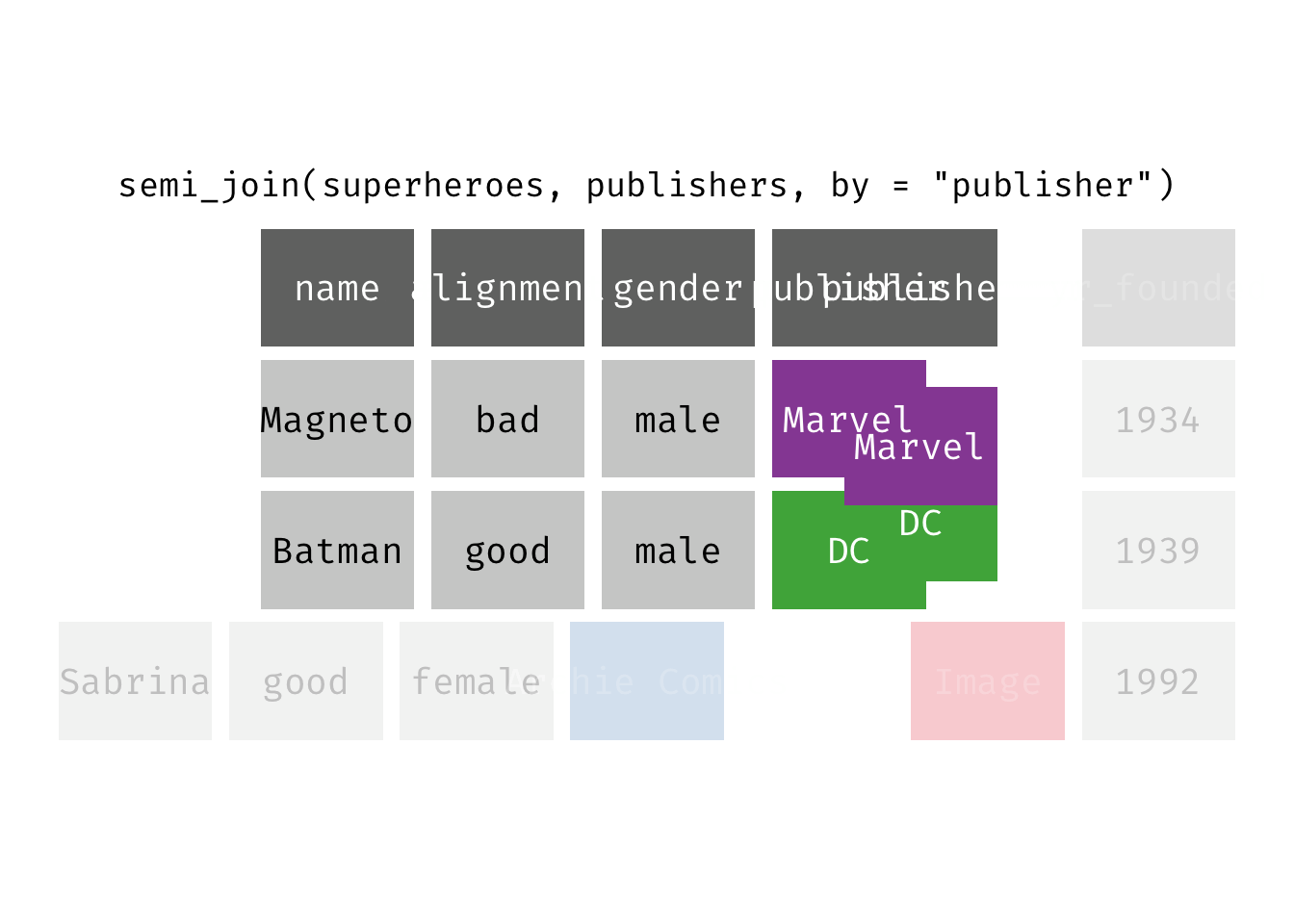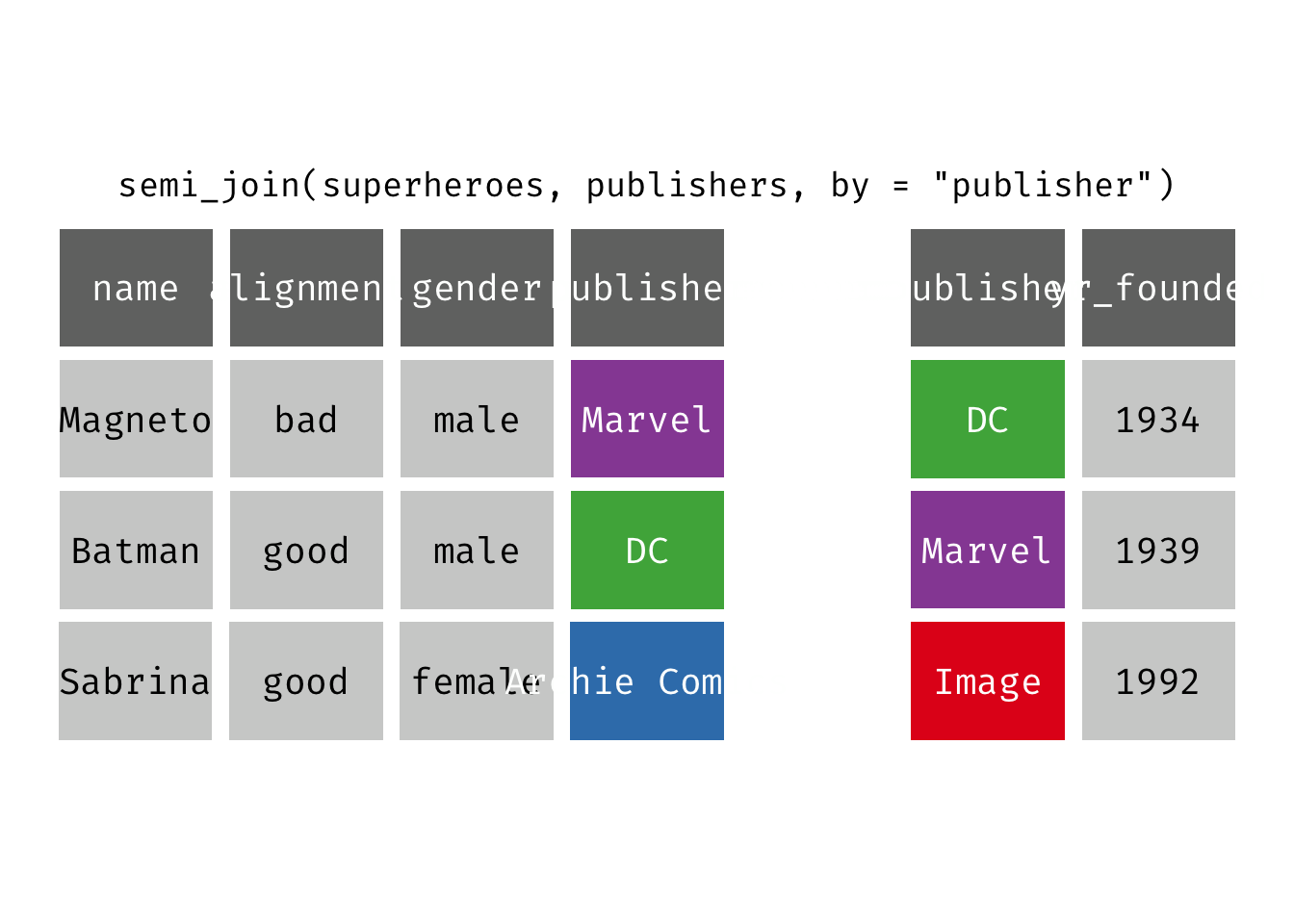
(sjsp <- semi_join(x = superheroes, y = publishers))
## Joining, by = "publisher"
## # A tibble: 2 × 4
## name alignment gender publisher
## <chr> <chr> <chr> <chr>
## 1 Magneto bad male Marvel
## 2 Batman good male DC
We get a similar result as with inner_join() but the join result contains only the variables originally found in x = superheroes. But note the row order has changed.
Anti join
anti_join(x, y): Return all rows from x where there are not matching values in y, keeping just columns from x. This is a filtering join.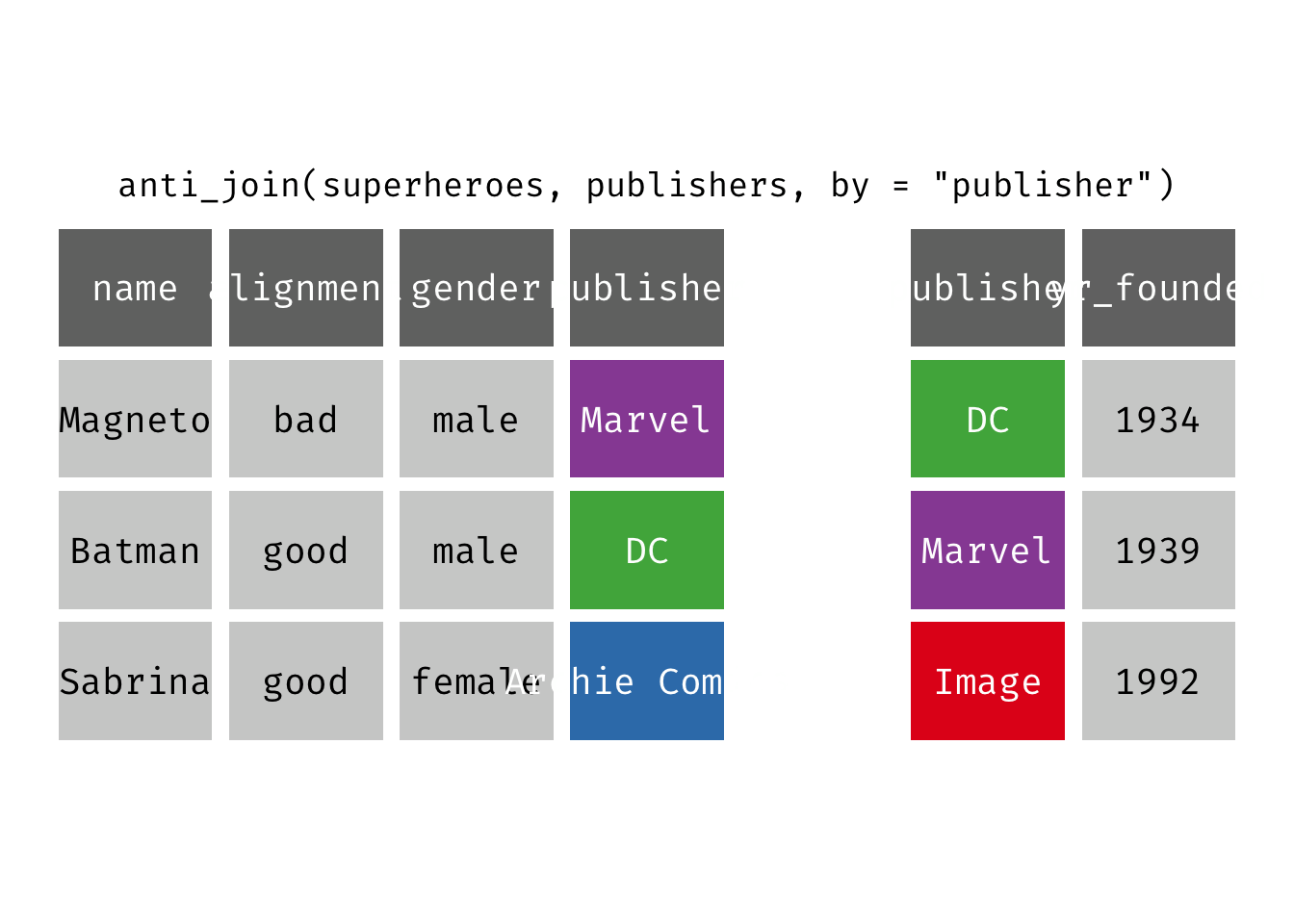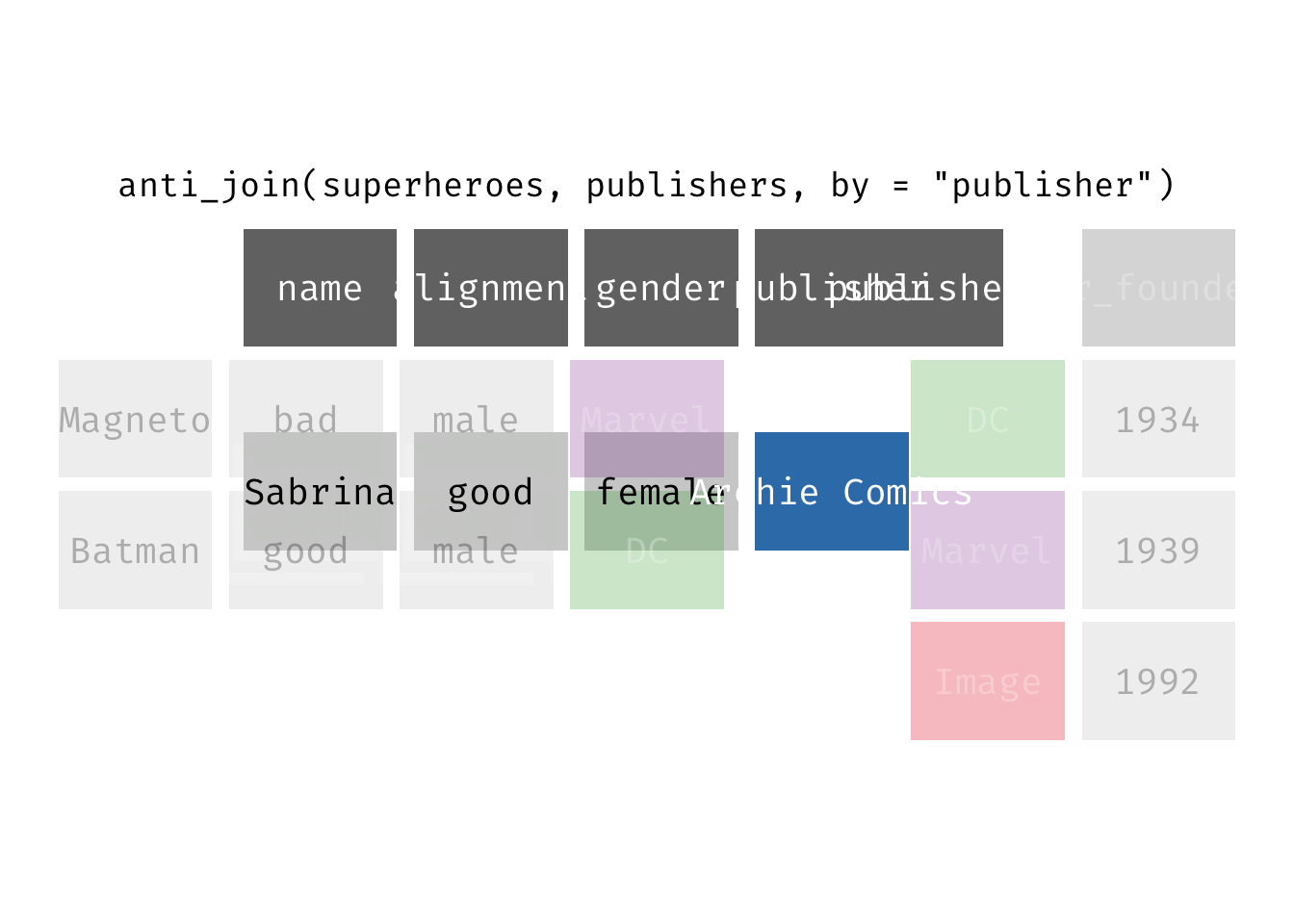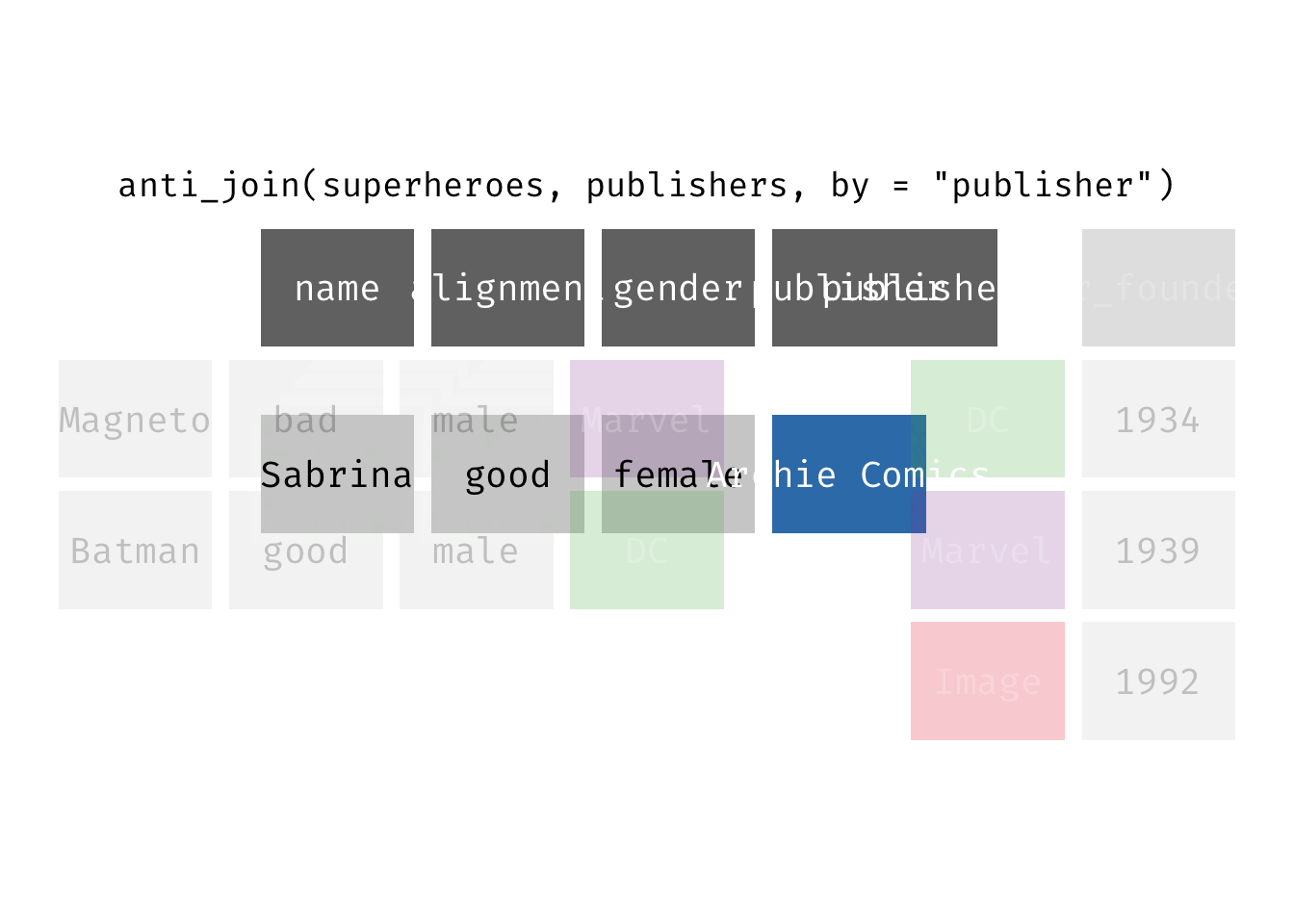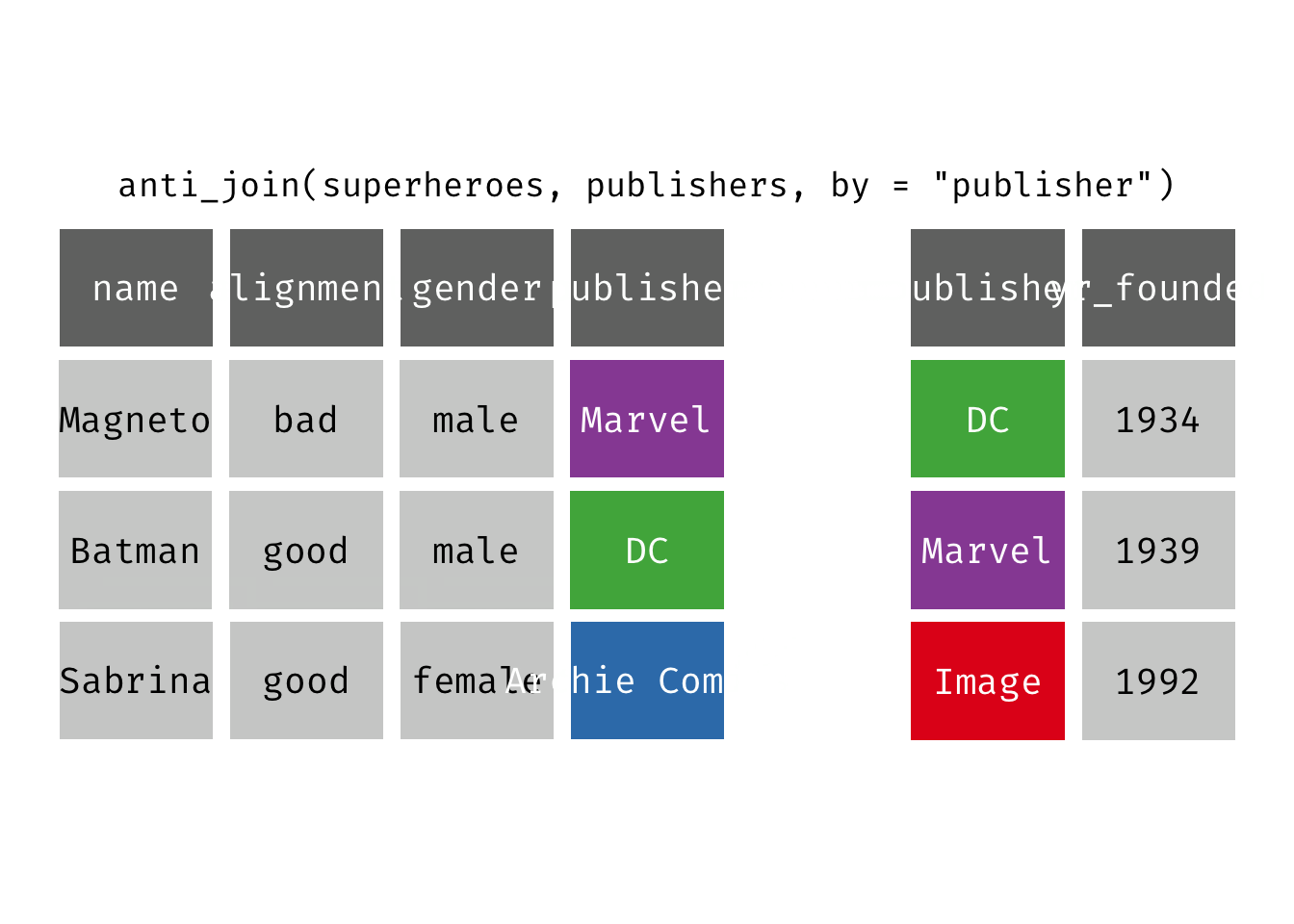
(ajsp <- anti_join(x = superheroes, y = publishers))
## Joining, by = "publisher"
## # A tibble: 1 × 4
## name alignment gender publisher
## <chr> <chr> <chr> <chr>
## 1 Sabrina good female Archie Comics
We keep only Sabrina now (and do not get yr_founded).
Acknowledgments
- This page is derived in part from “UBC STAT 545A and 547M”, licensed under the CC BY-NC 3.0 Creative Commons License.
Session Info
sessioninfo::session_info()
## ─ Session info ───────────────────────────────────────────────────────────────
## setting value
## version R version 4.2.1 (2022-06-23)
## os macOS Monterey 12.3
## system aarch64, darwin20
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz America/New_York
## date 2022-08-22
## pandoc 2.18 @ /Applications/RStudio.app/Contents/MacOS/quarto/bin/tools/ (via rmarkdown)
##
## ─ Packages ───────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## assertthat 0.2.1 2019-03-21 [2] CRAN (R 4.2.0)
## blogdown 1.10 2022-05-10 [2] CRAN (R 4.2.0)
## bookdown 0.27 2022-06-14 [2] CRAN (R 4.2.0)
## bslib 0.4.0 2022-07-16 [2] CRAN (R 4.2.0)
## cachem 1.0.6 2021-08-19 [2] CRAN (R 4.2.0)
## cli 3.3.0 2022-04-25 [2] CRAN (R 4.2.0)
## colorspace 2.0-3 2022-02-21 [2] CRAN (R 4.2.0)
## crayon 1.5.1 2022-03-26 [2] CRAN (R 4.2.0)
## DBI 1.1.3 2022-06-18 [2] CRAN (R 4.2.0)
## digest 0.6.29 2021-12-01 [2] CRAN (R 4.2.0)
## dplyr 1.0.9 2022-04-28 [2] CRAN (R 4.2.0)
## ellipsis 0.3.2 2021-04-29 [2] CRAN (R 4.2.0)
## evaluate 0.16 2022-08-09 [1] CRAN (R 4.2.1)
## fansi 1.0.3 2022-03-24 [2] CRAN (R 4.2.0)
## farver 2.1.1 2022-07-06 [2] CRAN (R 4.2.0)
## fastmap 1.1.0 2021-01-25 [2] CRAN (R 4.2.0)
## generics 0.1.3 2022-07-05 [2] CRAN (R 4.2.0)
## gganimate * 1.0.7 2020-10-15 [2] CRAN (R 4.2.0)
## ggplot2 * 3.3.6 2022-05-03 [2] CRAN (R 4.2.0)
## gifski 1.6.6-1 2022-04-05 [2] CRAN (R 4.2.0)
## glue 1.6.2 2022-02-24 [2] CRAN (R 4.2.0)
## gtable 0.3.0 2019-03-25 [2] CRAN (R 4.2.0)
## here 1.0.1 2020-12-13 [2] CRAN (R 4.2.0)
## hms 1.1.1 2021-09-26 [2] CRAN (R 4.2.0)
## htmltools 0.5.3 2022-07-18 [2] CRAN (R 4.2.0)
## jquerylib 0.1.4 2021-04-26 [2] CRAN (R 4.2.0)
## jsonlite 1.8.0 2022-02-22 [2] CRAN (R 4.2.0)
## knitr 1.39 2022-04-26 [2] CRAN (R 4.2.0)
## lifecycle 1.0.1 2021-09-24 [2] CRAN (R 4.2.0)
## magrittr 2.0.3 2022-03-30 [2] CRAN (R 4.2.0)
## munsell 0.5.0 2018-06-12 [2] CRAN (R 4.2.0)
## pillar 1.8.0 2022-07-18 [2] CRAN (R 4.2.0)
## pkgconfig 2.0.3 2019-09-22 [2] CRAN (R 4.2.0)
## prettyunits 1.1.1 2020-01-24 [2] CRAN (R 4.2.0)
## progress 1.2.2 2019-05-16 [2] CRAN (R 4.2.0)
## purrr 0.3.4 2020-04-17 [2] CRAN (R 4.2.0)
## R6 2.5.1 2021-08-19 [2] CRAN (R 4.2.0)
## Rcpp 1.0.9 2022-07-08 [2] CRAN (R 4.2.0)
## rlang 1.0.4 2022-07-12 [2] CRAN (R 4.2.0)
## rmarkdown 2.14 2022-04-25 [2] CRAN (R 4.2.0)
## rprojroot 2.0.3 2022-04-02 [2] CRAN (R 4.2.0)
## rstudioapi 0.13 2020-11-12 [2] CRAN (R 4.2.0)
## sass 0.4.2 2022-07-16 [2] CRAN (R 4.2.0)
## scales 1.2.0 2022-04-13 [2] CRAN (R 4.2.0)
## sessioninfo 1.2.2 2021-12-06 [2] CRAN (R 4.2.0)
## stringi 1.7.8 2022-07-11 [2] CRAN (R 4.2.0)
## stringr 1.4.0 2019-02-10 [2] CRAN (R 4.2.0)
## tibble 3.1.8 2022-07-22 [2] CRAN (R 4.2.0)
## tidyexplain * 0.0.1.9000 2022-08-16 [1] Github (gadenbuie/tidyexplain@7c9b6bf)
## tidyr 1.2.0 2022-02-01 [2] CRAN (R 4.2.0)
## tidyselect 1.1.2 2022-02-21 [2] CRAN (R 4.2.0)
## tweenr 1.0.2 2021-03-23 [2] CRAN (R 4.2.0)
## utf8 1.2.2 2021-07-24 [2] CRAN (R 4.2.0)
## vctrs 0.4.1 2022-04-13 [2] CRAN (R 4.2.0)
## withr 2.5.0 2022-03-03 [2] CRAN (R 4.2.0)
## xfun 0.31 2022-05-10 [1] CRAN (R 4.2.0)
## yaml 2.3.5 2022-02-21 [2] CRAN (R 4.2.0)
##
## [1] /Users/soltoffbc/Library/R/arm64/4.2/library
## [2] /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library
##
## ──────────────────────────────────────────────────────────────────────────────